Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Section Editors
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
BIOLOGY MOLECULAR / CLINICAL RESEARCH
The SP1/SNHG16/GLUT1 axis promotes prostate cancer proliferation and invasion by regulating glucose metabolism
1
Department of Urology, The First Affiliated Hospital and College of Clinical Medicine of Henan University of Science and Technology, Luoyang, Henan, China
2
Department of General Practice, The First Affiliated Hospital and College of Clinical Medicine of Henan University of Science and Technology, Luoyang, Henan, China
Submission date: 2024-01-30
Final revision date: 2024-07-02
Acceptance date: 2024-07-12
Online publication date: 2024-07-28
Corresponding author
Hua Huang
Department of Urology, The First Affiliated Hospital and College of Clinical Medicine of Henan University of Science and Technology, Luoyang, Henan 471000,, 471000, Luoyang, China
Department of Urology, The First Affiliated Hospital and College of Clinical Medicine of Henan University of Science and Technology, Luoyang, Henan 471000,, 471000, Luoyang, China
KEYWORDS
TOPICS
ABSTRACT
Introduction:
The SP1/SNHG16/GLUT1 axis is involved in diverse cancer-related processes. This study was designed to study the role of the SP1/SNHG16/GLUT1 axis in prostate cancer (PCa) via modulation of glucose metabolism.
Material and methods:
The expression profile of SNHG16 in PCa tissues was obtained from the online public database GEPIA (http://gepia.cancer-pku.cn/). Human prostate cancer cell lines, PC-3 and DU-145, were used in this study. Real-time qPCR was employed to determine the mRNA expression levels of SNHG16 and GLUT1. To assess cellular proliferation, CCK-8 assays were performed. Cellular invasion was evaluated using Transwell assays, and glycolysis was monitored through glucose uptake, lactate production, and ATP generation measurements.
Results:
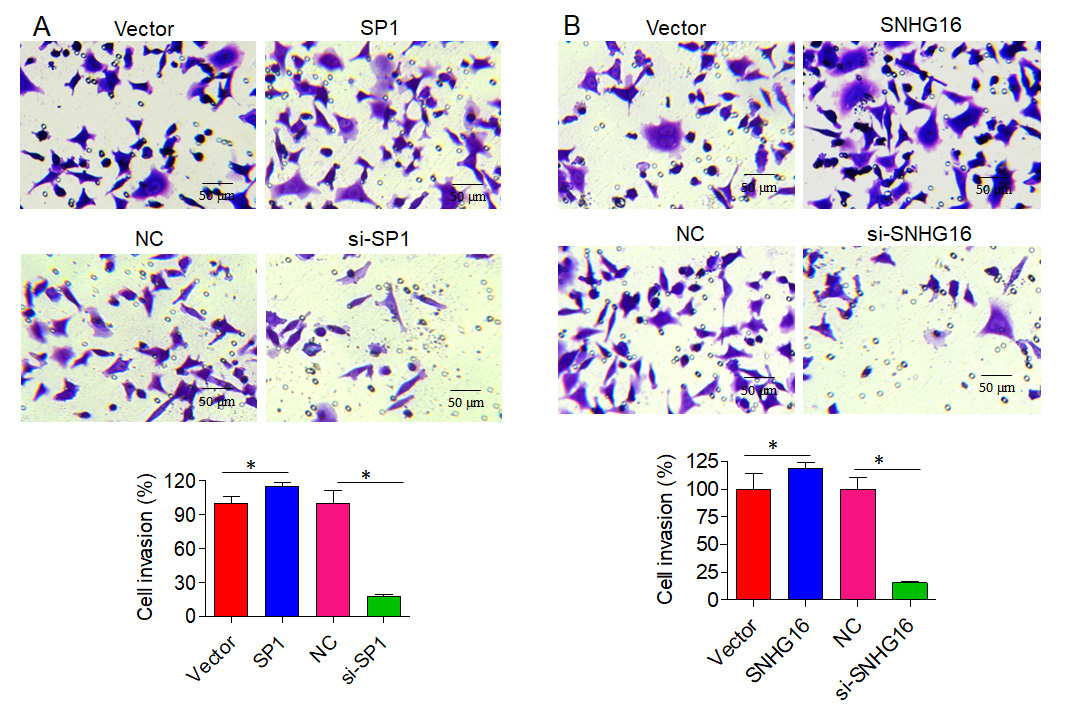
Analysis of GEPIA2 data revealed upregulation of SNHG16 in PCa tissues and a positive association between GLUT1 (SLC2A1) and SP1/SNHG16 expression in the correlation study. Consistently, SPI and SNHG16 were either overexpressed or knocked down in PCa cells to reveal the role of the SP1/SNHG16/GLUT1 axis. The results demonstrated that PC-3 and DU145 cell proliferation was promoted by the overexpression of either SPI or SNHG16. On the other hand, PC-3 and DU145 cell proliferation was reduced upon knockdown of SP1 or SNHG16. A real-time qPCR study revealed that GLUT1 mRNA was upregulated by SP1/SNHG16 overexpression and downregulated by SP1/SNHG16 knockdown. In PCa cells, overexpression of SNHG16/SP1 resulted in enhanced utilization of glucose, lactose, and ATP production, whereas SNHG16/SP1 knockdown had the reverse effect. Lastly, Transwell assay results showed that overexpression of SNHG16/SP1 promoted, while knockdown of SNHG16/SP1 inhibited, the invasiveness of PC-3 and DU145 PCa cells.
Conclusions:
Collectively, the evidence indicates that the SP1/SNHG16/GLUT1 axis regulates proliferation of PCa cells via the glycolytic route and thus may act as a therapeutic target for PCa treatment.
The SP1/SNHG16/GLUT1 axis is involved in diverse cancer-related processes. This study was designed to study the role of the SP1/SNHG16/GLUT1 axis in prostate cancer (PCa) via modulation of glucose metabolism.
Material and methods:
The expression profile of SNHG16 in PCa tissues was obtained from the online public database GEPIA (http://gepia.cancer-pku.cn/). Human prostate cancer cell lines, PC-3 and DU-145, were used in this study. Real-time qPCR was employed to determine the mRNA expression levels of SNHG16 and GLUT1. To assess cellular proliferation, CCK-8 assays were performed. Cellular invasion was evaluated using Transwell assays, and glycolysis was monitored through glucose uptake, lactate production, and ATP generation measurements.
Results:
Analysis of GEPIA2 data revealed upregulation of SNHG16 in PCa tissues and a positive association between GLUT1 (SLC2A1) and SP1/SNHG16 expression in the correlation study. Consistently, SPI and SNHG16 were either overexpressed or knocked down in PCa cells to reveal the role of the SP1/SNHG16/GLUT1 axis. The results demonstrated that PC-3 and DU145 cell proliferation was promoted by the overexpression of either SPI or SNHG16. On the other hand, PC-3 and DU145 cell proliferation was reduced upon knockdown of SP1 or SNHG16. A real-time qPCR study revealed that GLUT1 mRNA was upregulated by SP1/SNHG16 overexpression and downregulated by SP1/SNHG16 knockdown. In PCa cells, overexpression of SNHG16/SP1 resulted in enhanced utilization of glucose, lactose, and ATP production, whereas SNHG16/SP1 knockdown had the reverse effect. Lastly, Transwell assay results showed that overexpression of SNHG16/SP1 promoted, while knockdown of SNHG16/SP1 inhibited, the invasiveness of PC-3 and DU145 PCa cells.
Conclusions:
Collectively, the evidence indicates that the SP1/SNHG16/GLUT1 axis regulates proliferation of PCa cells via the glycolytic route and thus may act as a therapeutic target for PCa treatment.
REFERENCES (30)
1.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020; 70: 7-30.
2.
Culp MB, Soerjomataram I, Efstathiou JA, et al. Recent global patterns in prostate cancer incidence and mortality rates. Eur Urol 2020; 77: 38-52.
3.
Litwin MS, Tan HJ. The diagnosis and treatment of prostate cancer: a review. JAMA 2017; 317: 2532-42.
4.
Qiu H, Cao S, Xu R. Cancer incidence, mortality, and burden in China: a time-trend analysis and comparison with the United States and United Kingdom based on the global epidemiological data released in 2020. Cancer Commun (Lond) 2021; 41: 1037-48.
5.
Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab 2016; 23: 27-47.
6.
Carvalho TM, Cardoso HJ, Figueira MI, et al. The peculiarities of cancer cell metabolism: a route to metastasization and a target for therapy. Eur J Med Chem 2019; 171: 343-63.
7.
Srihari S, Kwong R, Tran K, et al. Metabolic deregulation in prostate cancer. Mol Omics 2018; 14: 320-9.
8.
Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 2012; 21: 297-8.
9.
Jakoby P, Schmidt E, Ruminot I et al. Higher transport and metabolism of glucose in astrocytes compared with neurons: a multiphoton study of hippocampal and cerebellar tissue slices. Cereb Cortex 2014; 24: 222-31.
10.
Quincozes-Santos A, Bobermin LD, de Assis AM, et al. Fluctuations in glucose levels induce glial toxicity with glutamatergic, oxidative and inflammatory implications. Biochim Biophys Acta Mol Basis Dis 2017; 1863: 1-14.
11.
Mirzaei S, Paskeh MDA, Okina E, et al. Molecular landscape of LncRNAs in prostate cancer: a focus on pathways and therapeutic targets for intervention. J Exp Clin Cancer Res 2022; 41: 214.
12.
Han C, Yang Y, Guo L, Guan Q, Ruan S. The expression of long non-coding RNA HOTAIR in advanced hepatocellular carcinoma and its prognostic correlation with sunitinib therapy. Arch Med Sci 2021; 18: 71-8.
13.
Wang J, Li J. LncRNA MIR155HG functions as a ceRNA for inhibition of lung adenocarcinoma growth and prediction of prognosis. Arch Med Sci 2021; 20: 539-56.
14.
Yang M, Wei W. SNHG16: a novel long-non-coding RNA in human cancers. Onco Targets Ther 2019; 12: 11679-90.
15.
Gong CY, Tang R, Nan W, et al. Role of SNHG16 in human cancer. Clin Chim Acta 2020; 503: 175-80.
16.
Vizcaíno C, Mansilla S, Portugal J. Sp1 transcription factor: a long-standing target in cancer chemotherapy. Pharmacol Ther 2015; 152: 111-24.
17.
Hasegawa Y, Struhl K. Different SP1 binding dynamics at individual genomic loci in human cells. Proc Natl Acad Sci USA 2021; 118: e2113579118.
18.
Yin J, Shi Z, Wei W, et al. MiR-181b suppress glioblastoma multiforme growth through inhibition of SP1-mediated glucose metabolism. Cancer Cell Int 2020; 20: 69.
19.
Beishline K, Azizkhan-Clifford J. Sp1 and the ‘hallmarks of cancer’. FEBS J 2015; 282: 224-58.
20.
Feng W, Cui G, Tang CW, et al. Role of glucose metabolism related gene GLUT1 in the occurrence and prognosis of colorectal cancer. Oncotarget 2017; 8: 56850.
21.
Nakas Abate-Shen C, Shen MM. Molecular genetics of prostate cancer. Gen Develop 2000; 14: 2410-34.
23.
Yang XS, Wang GX, Luo L. Long non-coding RNA SNHG16 promotes cell growth and metastasis in ovarian cancer. Eur Rev Med Pharma Sci 2018; 22: 616-22.
24.
Lian D, Amin B, Du D, et al. Enhanced expression of the long non-coding RNA SNHG16 contributes to gastric cancer progression and metastasis. Cancer Biomarkers 2018; 21: 151-60.
25.
Shao M, Yu Z, Zou J. LncRNA-SNHG16 silencing inhibits prostate carcinoma cell growth, downregulate GLUT1 expression and reduce glucose uptake. Cancer Manag Res 2020; 12: 1751-7.
26.
Liu Y, Zuckier LS, Ghesani NV. Dominant uptake of fatty acid over glucose by prostate cells: a potential new diagnostic and therapeutic approach. Anticancer Res 2010; 30: 369-74.
27.
Sadeghi RN, Karami-Tehrani F, Salami S. Targeting prostate cancer cell metabolism: impact of hexokinase and CPT-1 enzymes. Tumour Biol 2015; 36: 2893-905.
28.
Arfin S, Jha NK, Jha SK, et al. Oxidative stress in cancer cell metabolism. Antioxidants (Basel) 2021; 10: 642.
29.
Xie X, Xu X, Sun C, et al. Long intergenic noncoding RNA SNHG16 interacts with miR-195 to promote proliferation, invasion and tumorigenesis in hepatocellular carcinoma. Exp Cell Res 2019; 383: 111501.
30.
Su P, Mu S, Wang Z. Long noncoding RNA SNHG16 promotes osteosarcoma cells migration and invasion via sponging miRNA-340. DNA Cell Biol 2019; 38: 170-5.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.