Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
NEUROLOGY / BASIC RESEARCH
Single cell sequencing reveals heterogenicity of differential gene expression and altered interactome in post-ischemic mouse brain cells
1
Department of Neurology, The First Affiliated Hospital of Shandong First Medical University, Jinan City, Shandong Province, 250014, China
2
Department of Neurology, Affiliated Hospital of Shandong University of Traditional Chinese Medicine, Jinan City, Shandong Province, 250014, China
3
Department of Gastroenterology, The First Affiliated Hospital of Shandong First Medical University, Jinan City, Shandong Province, 250014, China
These authors had equal contribution to this work
Submission date: 2024-03-05
Final revision date: 2024-05-25
Acceptance date: 2024-06-14
Online publication date: 2024-06-21
Corresponding author
Jinping Zhang
Department of Neurology, Affiliated Hospital of Shandong University of Traditional Chinese Medicine, No. 16369#, Jingshi Road, Jinan City, Shandong Province, 250014, China, China
Department of Neurology, Affiliated Hospital of Shandong University of Traditional Chinese Medicine, No. 16369#, Jingshi Road, Jinan City, Shandong Province, 250014, China, China
KEYWORDS
TOPICS
ABSTRACT
Introduction:
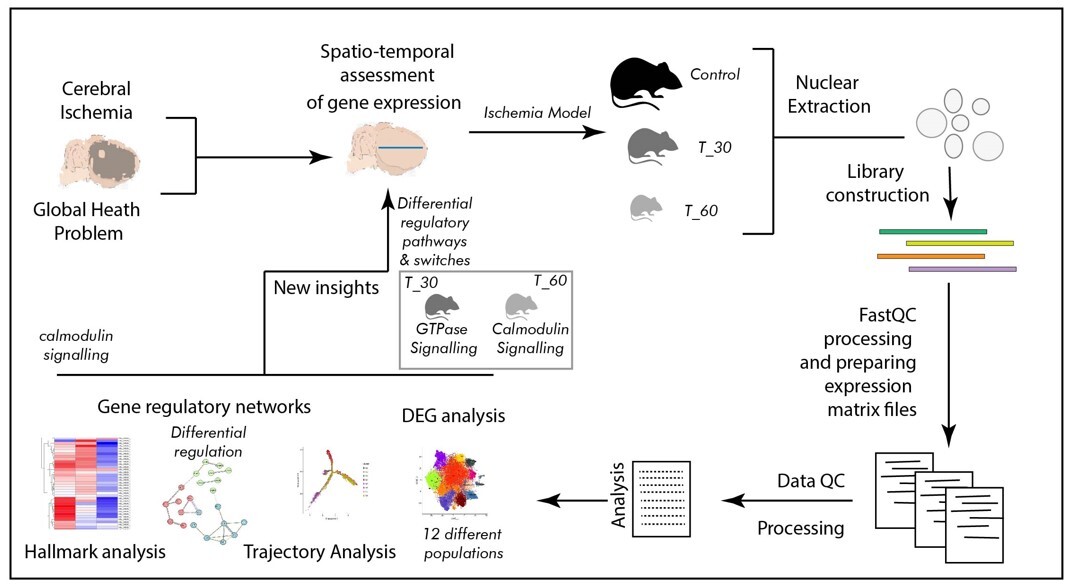
Ischemia, resulting from reduced blood supply, poses a critical health challenge. It is known to be caused by arterial constriction or blockages and triggers oxygen and nutrient deprivation, impacting multiple body systems and leading to a multitude of diseases and associated health conditions. Due to its multifaceted association with several diseases, ischemia is a subject of interest in many clinical studies. Over several decades of information on ischemia and related molecular changes have provided valuable insight into its pathophysiological outcomes. However, the scarcity of molecular studies, especially genomic inquiries employing spatial and temporal segregation, remains mostly unaddressed. The emerging field of single-cell genomics offers promising solutions to such inquiries. Therefore, we performed our study by utilizing a single-cell genomics approach, employing a mouse brain model of hypoxia at two distinct time points (30 min and 60 min of exposure with hypoxia) to delineate cellular trajectories, ontology, and the clustering of expression-based patterns in a cell-specific manner.
Material and methods:
In the present study we developed a mouse model of hypoxia, which was established using the thread-plug method. The experimental groups were subjected to hypoxia for 30 min (T_30) and 60 min (T_60), while the sham surgery group was used as a control. Following excision of the cerebral cortex, nuclear isolation and library construction were performed before conducting spatio-temporal analysis of cortical cells. Comprehensive data analysis encompassed differential gene expression analysis, trajectory analysis, examination of gene regulatory networks, and hallmark analysis.
Results:
The primary outcome of the single cell genomics analysis emerged as clustering of 12 distinct cell populations suggesting contrasting transcriptomic profiles. Furthermore, spatio-temporal distinction in cell signaling was identified as a switch from Ras GTPase signaling to calmodulin and calcium dependent signaling between two levels of ischemia. The most dynamic regions in terms of transcription were distal axons and growth cones in the T_30 group, and cell edges and the post-synaptic area in the T_60 group. Also, the synaptic vesicle cycle is likely to be involved in such transcription switching.
Conclusions:
Our study employing a single-cell genomics approach provides valuable insights into the cellular dynamics during hypoxia exposure. The identified cell populations and associated molecular pathways offer potential targets for further research and development of targeted therapies in addressing the complex challenges posed by ischemia.
Ischemia, resulting from reduced blood supply, poses a critical health challenge. It is known to be caused by arterial constriction or blockages and triggers oxygen and nutrient deprivation, impacting multiple body systems and leading to a multitude of diseases and associated health conditions. Due to its multifaceted association with several diseases, ischemia is a subject of interest in many clinical studies. Over several decades of information on ischemia and related molecular changes have provided valuable insight into its pathophysiological outcomes. However, the scarcity of molecular studies, especially genomic inquiries employing spatial and temporal segregation, remains mostly unaddressed. The emerging field of single-cell genomics offers promising solutions to such inquiries. Therefore, we performed our study by utilizing a single-cell genomics approach, employing a mouse brain model of hypoxia at two distinct time points (30 min and 60 min of exposure with hypoxia) to delineate cellular trajectories, ontology, and the clustering of expression-based patterns in a cell-specific manner.
Material and methods:
In the present study we developed a mouse model of hypoxia, which was established using the thread-plug method. The experimental groups were subjected to hypoxia for 30 min (T_30) and 60 min (T_60), while the sham surgery group was used as a control. Following excision of the cerebral cortex, nuclear isolation and library construction were performed before conducting spatio-temporal analysis of cortical cells. Comprehensive data analysis encompassed differential gene expression analysis, trajectory analysis, examination of gene regulatory networks, and hallmark analysis.
Results:
The primary outcome of the single cell genomics analysis emerged as clustering of 12 distinct cell populations suggesting contrasting transcriptomic profiles. Furthermore, spatio-temporal distinction in cell signaling was identified as a switch from Ras GTPase signaling to calmodulin and calcium dependent signaling between two levels of ischemia. The most dynamic regions in terms of transcription were distal axons and growth cones in the T_30 group, and cell edges and the post-synaptic area in the T_60 group. Also, the synaptic vesicle cycle is likely to be involved in such transcription switching.
Conclusions:
Our study employing a single-cell genomics approach provides valuable insights into the cellular dynamics during hypoxia exposure. The identified cell populations and associated molecular pathways offer potential targets for further research and development of targeted therapies in addressing the complex challenges posed by ischemia.
REFERENCES (41)
2.
Kalogeris T, Baines CP, Krenz M, Korthuis RJ. Cell biology of ischemia/reperfusion injury. Int Rev Cell Mol Biol 2012; 298: 229-317.
3.
Price AJ, Wright FL, Green J, et al. Differences in risk factors for 3 types of stroke: UK prospective study and meta-analyses. Neurology 2018; 90: e298-e306.
4.
Soler EP, Ruiz VC. Epidemiology and risk factors of cerebral ischemia and ischemic heart diseases: similarities and differences. Curr Cardiol Rev 2010; 6: 138-49.
6.
Feigin VL, Lawes CM, Bennett DA, Anderson CS. Stroke epidemiology: a review of population-based studies of incidence, prevalence, and case-fatality in the late 20th century. Lancet Neurol 2003; 2: 43-53.
7.
Tu WJ, Zhao Z, Yin P, et al. Estimated burden of stroke in China in 2020. JAMA Netw Open 2023; 6: e231455.
8.
Wang W, Jiang B, Sun H, et al. Prevalence, incidence, and mortality of stroke in China: results from a nationwide population-based survey of 480 687 adults. Circulation 2017; 135: 759-71.
9.
Rubin MN, Barrett KM. What to do with wake-up stroke. Neurohospitalist 2015; 5: 161-72.
11.
Trivedi JK. Cognitive deficits in psychiatric disorders: current status. Indian J Psychiatry 2006; 48: 10-20.
12.
Imran R, Mohamed GA, Nahab F. Acute reperfusion therapies for acute ischemic stroke. J Clin Med 2021; 10: 3677.
13.
Okumura E, Tsurukiri J, Ota T, et al. Outcomes of endovascular thrombectomy performed 6-24 h after acute stroke from extracranial internal carotid artery occlusion. Neurol Med Chir (Tokyo) 2019; 59: 337-43.
14.
Winnige P, Vysoky R, Dosbaba F, Batalik L. Cardiac rehabilitation and its essential role in the secondary prevention of cardiovascular diseases. World J Clin Cases 2021; 9: 1761-84.
15.
Herpich F, Rincon F. Management of acute ischemic stroke. Crit Care Med 2020; 48: 1654-63.
16.
Campbell BCV, De Silva DA, Macleod MR, et al. Ischaemic stroke. Nat Rev Dis Primers 2019; 5: 70.
17.
Safdieh JE, Govindarajan R, Gelb DJ, Odia Y, Soni M. Core curriculum guidelines for a required clinical neurology experience Neurology 2019; 92: 619-626. Erratum in: Neurology 2019; 93: 135.
18.
de Sena Brandine G, Smith AD. Falco: high-speed FastQC emulation for quality control of sequencing data. F1000Res 2019; 8: 1874.
19.
Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018; 34: i884-90.
20.
Kechin A, Boyarskikh U, Kel A, Filipenko M. cutPrimers: A New Tool for Accurate Cutting of Primers from Reads of Targeted Next Generation Sequencing. J Comput Biol 2017; 24: 1138-43.
21.
Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29: 15-21.
22.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014; 30: 923-30.
23.
Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 2019; 16: 1289-96.
24.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012; 16: 284-7.
25.
Aibar S, González-Blas CB, Moerman T, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 2017; 14: 1083-6.
26.
Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat Methods 2017; 14: 309-15.
27.
Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 2020; 15: 1484-506.
28.
Porteous MK, Fritz JS. Hypoxemia in a patient with pulmonary arterial hypertension: getting to the heart of the matter. Ann Am Thorac Soc 2014; 11: 836-40.
29.
Tang F, Barbacioru C, Wang Y, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 2009; 6: 377-82.
30.
Zhang Y, Zhang B, Lv C, et al. Single-cell RNA sequencing identifies critical transcription factors of tumor cell invasion induced by hypoxia microenvironment in glioblastoma. Theranostics 2023; 13: 3744-60.
31.
Prass K, Scharff A, Ruscher K, et al. Hypoxia-induced stroke tolerance in the mouse is mediated by erythropoietin. Stroke 2003; 34: 1981-6.
32.
Fluri F, Schuhmann MK, Kleinschnitz C. Animal models of ischemic stroke and their application in clinical research. Drug Des Devel Ther 2015; 9: 3445-54.
33.
Thomas S, Manivannan S, Garg V, Lilly B. Single-cell RNA sequencing reveals novel genes regulated by hypoxia in the lung vasculature. J Vasc Res 2022; 59: 163-75.
34.
Guo K, Luo J, Feng D, et al. Single-cell RNA sequencing with combined use of bulk RNA sequencing to reveal cell heterogeneity and molecular changes at acute stage of ischemic stroke in mouse cortex penumbra area. Front Cell Dev Biol 2021; 9: 624711.
35.
Ma H, Zhou Y, Li Z, et al. Single-cell RNA-sequencing analyses revealed heterogeneity and dynamic changes of metabolic pathways in astrocytes at the acute phase of ischemic stroke. Oxid Med Cell Longev 2022; 2022: 1817721.
36.
Mao XG, Xue XY, Wang L, et al. CDH5 is specifically activated in glioblastoma stemlike cells and contributes to vasculogenic mimicry induced by hypoxia. Neuro Oncol 2013; 15: 865-79.
37.
Conte C, Riant E, Toutain C, et al. FGF2 translationally induced by hypoxia is involved in negative and positive feedback loops with HIF-1alpha. PLoS One 2008; 3: e3078.
38.
Sakai D, Sugawara T, Kurokawa T, et al. Hif1α-dependent hypoxia signaling contributes to the survival of deep-layer neurons and cortex formation in a mouse model. Mol Brain 2022; 15: 28.
39.
Issac MSM, Yousef E, Tahir MR, Gaboury LA. MCM2, MCM4, and MCM6 in breast cancer: clinical utility in diagnosis and prognosis. Neoplasia 2019; 21: 1015-35.
40.
Zhu S, Tang J, Lan L, Su F. Inhibition of miR-34a ameliorates cerebral ischemia/reperfusion injury by targeting brain-derived neurotrophic factor. Arch Med Sci 2021. doi: 10.5114/aoms/143303.
41.
Duan Q, Sun W, Yuan H, Mu X. MicroRNA-135b-5p prevents oxygen-glucose deprivation and reoxygenation-induced neuronal injury through regulation of the GSK-3β/Nrf2/ARE signaling pathway. Arch Med Sci 2018; 14: 735-44.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.