Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Section Editors
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
CLINICAL RESEARCH
Risk stratification of soft tissue sarcoma based on activity of prognostic molecules associated with unpolarized macrophages
1
Department of Anatomy, Institute of Neuroscience, Basic Medical College. Chongqing Medical University, Chongqing, China
2
Department of Plastic Surgery, Central Hospital Affiliated to Chongqing University of Technology, Chongqing, China
3
Department of Oncology, Jinshazhou Hospital of Guangzhou University of Chinese Medicine, Guangzhou, China
4
Department of Anatomy, Institute of Neuroscience, Basic Medical College, Chongqing Medical University, Chongqing, China
5
Department of Radiology, Central Hospital Affiliated to Chongqing University of Technology, Chongqing, China
These authors had equal contribution to this work
Submission date: 2024-08-31
Final revision date: 2024-11-30
Acceptance date: 2025-01-01
Online publication date: 2025-02-13
Corresponding author
Guiqiong He
Department of Anatomy Institute of Neuroscience Basic Medical College Chongqing Medical University Chongqing, China
Department of Anatomy Institute of Neuroscience Basic Medical College Chongqing Medical University Chongqing, China
Hong Lu
Department of Radiology Central Hospital Affiliated to Chongqing University of Technology Chongqing, China
Department of Radiology Central Hospital Affiliated to Chongqing University of Technology Chongqing, China
KEYWORDS
TOPICS
ABSTRACT
Introduction:
Soft tissue sarcomas (STS), representing 80% of sarcomas, are a rare and diverse group of neoplasms with a dire prognosis. Macrophages and their subtypes play an essential role with diverse outcomes in the tumor microenvironment (TME) of cancers, including sarcomas. The aim of this study was to investigate the role of macrophages in the development and prognosis of sarcoma patients.
Material and methods:
Transcriptomic data from 5 sarcoma cohorts including 581 patients and transcripts of 56,752 single cells from 6 sarcoma patients were retrieved from public databases and analyzed. The infiltration of immune cells in the TME was evaluated with the CIBERSORT algorithm. Kaplan-Meier estimation with the log-rank test and Cox regression hazards models were adopted for evaluation of prognostic impacts.
Results:
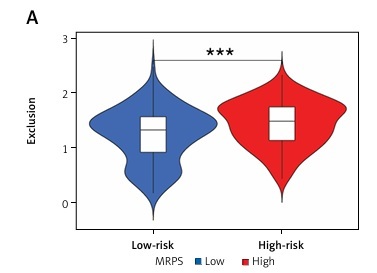
Deconvolution of 22 types of immune cells via the CIBERSORT algorithm revealed macrophages as a prominent component of the TME of sarcoma patients. Of these, M0 was associated with worst prognosis. A six-gene prognostic signature, termed MRPS, was developed that was significantly positively correlated with M0 macrophages. The MRPS-stratified high-risk subgroup showed abundance of M0 macrophages, indicating inhibition of macrophage polarization, specifically the classically activated pro-inflammatory M1 phenotype. Moreover, enrichment of oncogenic pathways and glycolysis and high frequency of mutations were evident. The robustness of the MRPS as a predictive biomarker was validated in external soft-tissue sarcoma patient datasets. A nomogram based on MRPS was developed as a potentially accurate and practical predictive tool for identifying high-risk sarcoma patients with lower survival probabilities. Furthermore, the MRPS signature exhibited reliable predictive capabilities for immunotherapy response, suggesting its potential to enhance the effectiveness of personalized immunotherapy in sarcoma patients.
Conclusions:
MRPS represents a robust biomarker for predicting outcomes and response to therapy in soft-tissue sarcoma patients.
Soft tissue sarcomas (STS), representing 80% of sarcomas, are a rare and diverse group of neoplasms with a dire prognosis. Macrophages and their subtypes play an essential role with diverse outcomes in the tumor microenvironment (TME) of cancers, including sarcomas. The aim of this study was to investigate the role of macrophages in the development and prognosis of sarcoma patients.
Material and methods:
Transcriptomic data from 5 sarcoma cohorts including 581 patients and transcripts of 56,752 single cells from 6 sarcoma patients were retrieved from public databases and analyzed. The infiltration of immune cells in the TME was evaluated with the CIBERSORT algorithm. Kaplan-Meier estimation with the log-rank test and Cox regression hazards models were adopted for evaluation of prognostic impacts.
Results:
Deconvolution of 22 types of immune cells via the CIBERSORT algorithm revealed macrophages as a prominent component of the TME of sarcoma patients. Of these, M0 was associated with worst prognosis. A six-gene prognostic signature, termed MRPS, was developed that was significantly positively correlated with M0 macrophages. The MRPS-stratified high-risk subgroup showed abundance of M0 macrophages, indicating inhibition of macrophage polarization, specifically the classically activated pro-inflammatory M1 phenotype. Moreover, enrichment of oncogenic pathways and glycolysis and high frequency of mutations were evident. The robustness of the MRPS as a predictive biomarker was validated in external soft-tissue sarcoma patient datasets. A nomogram based on MRPS was developed as a potentially accurate and practical predictive tool for identifying high-risk sarcoma patients with lower survival probabilities. Furthermore, the MRPS signature exhibited reliable predictive capabilities for immunotherapy response, suggesting its potential to enhance the effectiveness of personalized immunotherapy in sarcoma patients.
Conclusions:
MRPS represents a robust biomarker for predicting outcomes and response to therapy in soft-tissue sarcoma patients.
REFERENCES (67)
1.
Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F, editors. WHO Classification of Tumours of Soft Tissue and Bone. 4th ed. Lyon, France: IARC Press, International Agency for Research on Cancer; 2013.
2.
Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin 2023; 73: 17-48.
3.
Damerell V, Pepper MS, Prince S. Molecular mechanisms underpinning sarcomas and implications for current and future therapy. Signal Transduct Target Ther 2021; 6: 246.
4.
von Mehren M, Kane JM, Agulnik M, et al. Soft Tissue Sarcoma, Version 2.2022, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2022; 20: 815-33.
5.
Bleloch JS, Ballim RD, Kimani S, et al. Managing sarcoma: where have we come from and where are we going? Ther Adv Med Oncol 2017; 9: 637-59.
6.
Meyer M, Seetharam M. First-line therapy for metastatic soft tissue sarcoma. Curr Treat Options Oncol 2019; 20: 6.
7.
Carbonnaux M, Brahmi M, Schiffler C, et al. Very long-term survivors among patients with metastatic soft tissue sarcoma. Cancer Med 2019; 8: 1368-78.
8.
Wang Q, Shao X, Zhang Y, et al. Role of tumor microenvironment in cancer progression and therapeutic strategy. Cancer Med 2023; 12: 11149-65.
9.
Pandya PH, Murray ME, Pollok KE, Renbarger JL. The immune system in cancer pathogenesis: potential therapeutic approaches. J Immunol Res 2016; 2016: 4273943.
10.
Ren H, Bazhin AV, Pretzsch E, et al. A novel immune-related gene signature predicting survival in sarcoma patients. Mol Ther Oncolytics 2022; 24: 114-26.
11.
Weng W, Yu L, Li Z, et al. The immune subtypes and landscape of sarcomas. BMC Immunol 2022; 23: 46.
12.
Petitprez F, de Reyniès A, Keung EZ, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 2020; 577: 556-60.
13.
Sun D, Wang J, Han Y, et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res 2021; 49: D1420-d30.
14.
Wang C, Sun D, Huang X, et al. Integrative analyses of single- cell transcriptome and regulome using MAESTRO. Genome Biol 2020; 21: 198.
15.
Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013; 4: 2612.
16.
Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015; 12: 453-7.
17.
Jiang P, Gu S, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 2018; 24: 1550-8.
18.
Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making 2006; 26: 565-74.
19.
Cahlon O, Brennan MF, Jia X, Qin LX, Singer S, Alektiar KM. A postoperative nomogram for local recurrence risk in extremity soft tissue sarcomas after limb-sparing surgery without adjuvant radiation. Ann Surg 2012; 255: 343-7.
20.
Callegaro D, Miceli R, Bonvalot S, et al. Development and external validation of two nomograms to predict overall survival and occurrence of distant metastases in adults after surgical resection of localised soft-tissue sarcomas of the extremities: a retrospective analysis. Lancet Oncol 2016; 17: 671-80.
21.
Ren H, Bazhin AV, Pretzsch E, et al. A novel immune-related gene signature predicting survival in sarcoma patients. Mol Ther Oncol 2022; 24: 114-26.
22.
Xiao B, Liu L, Li A, et al. Identification and verification of immune-related gene prognostic signature based on ssGSEA for osteosarcoma. Front Oncol 2020; 10: 607622.
23.
Bui NQ, Przybyl J, Trabucco SE, et al. A clinico-genomic analysis of soft tissue sarcoma patients reveals CDKN2A deletion as a biomarker for poor prognosis. Clin Sarcoma Res 2019; 9: 12.
24.
Darmusey L, Pérot G, Thébault N, et al. ATRX alteration contributes to tumor growth and immune escape in pleomorphic sarcomas. Cancers 2021; 13: 2151.
25.
Yang H, Xiong F, Qi R, et al. LAPTM4B-35 is a novel prognostic factor of hepatocellular carcinoma. J Surg Oncol 2010; 101: 363-9.
26.
Wang L, Meng Y, Zhang QY. LAPTM4B is a novel diagnostic and prognostic marker for lung adenocarcinoma and associated with mutant EGFR. BMC Cancer 2019; 19: 293.
27.
Xiao M, Jia S, Wang H, Wang J, Huang Y, Li Z. Overexpression of LAPTM4B: an independent prognostic marker in breast cancer. J Cancer Res Clin Oncol 2013; 139: 661-7.
28.
Kang Y, Yin M, Jiang W, et al. Overexpression of LAPTM4B-35 is associated with poor prognosis in colorectal carcinoma. Am J Surg 2012; 204: 677-83.
29.
Zhang H, Tian B, Yu H, Yao H, Gao Z. LAPTM4B-35 protein as a potential therapeutic target in gastric cancer. Tumor Biol 2014; 35: 12737-42.
30.
Yang Z, Senninger N, Flammang I, Ye Q, Dhayat SA. Clinical impact of circulating LAPTM4B-35 in pancreatic ductal adenocarcinoma. J Cancer Res Clin Oncol 2019; 145: 1165-78.
31.
Meng F, Luo C, Hu Y, et al. Overexpression of LAPTM4B-35 in cervical carcinoma: a clinicopathologic study. Int J Gynecol Pathol 2010; 29: 587-93.
32.
Yin M, Li C, Li X, et al. Over-expression of LAPTM4B is associated with poor prognosis and chemotherapy resistance in stages III and IV epithelial ovarian cancer. J Surg Oncol 2011; 104: 29-36.
33.
Zhang H, Wei Q, Liu R, et al. Overexpression of LAPTM4B-35: a novel marker of poor prognosis of prostate cancer. PLoS One 2014; 9: e91069.
34.
Wang ZX, Guo MY, Ren J, Li GS, Sun XG. Identification of lysosome-associated protein transmembrane-4 as a novel therapeutic target for osteosarcoma treatment. Orthop Surg 2020; 12: 1253-60.
35.
Yang H, Xiong F, Wei X, Yang Y, McNutt MA, Zhou R. Overexpression of LAPTM4B-35 promotes growth and metastasis of hepatocellular carcinoma in vitro and in vivo. Cancer Letters 2010; 294: 236-44.
36.
Meng F, Chen X, Song H, Lou G. LAPTM4B down regulation inhibits the proliferation, invasion and angiogenesis of hela cells in vitro. Cell Physiol Biochem 2015; 37: 890-900.
37.
Liu M, Yan R, Wang J, Yao Z, Fan X, Zhou K. LAPTM4B-35 promotes cancer cell migration via stimulating integrin beta1 recycling and focal adhesion dynamics. Cancer Sci 2022; 113: 2022-33.
38.
Li Y, Zou L, Li Q, et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat Med 2010; 16: 214-8.
39.
Li Y, Zhang Q, Tian R, et al. Lysosomal transmembrane protein LAPTM4B promotes autophagy and tolerance to metabolic stress in cancer cells. Cancer Res 2011; 71: 7481-9.
40.
Woo IS, Eun SY, Jang HS, et al. Identification of ADP-ribosylation factor 4 as a suppressor of N-(4-hydroxyphenyl)retinamide-induced cell death. Cancer Lett 2009; 276: 53-60.
41.
Wu Q, Ren X, Zhang Y, et al. MiR-221-3p targets ARF4 and inhibits the proliferation and migration of epithelial ovarian cancer cells. Biochem Biophys Res Commun 2018; 497: 1162-70.
42.
Bidkhori G, Narimani Z, Hosseini Ashtiani S, Moeini A, Nowzari-Dalini A, Masoudi-Nejad A. Reconstruction of an integrated genome-scale co-expression network reveals key modules involved in lung adenocarcinoma. PLoS One 2013; 8: e67552.
43.
Jang SY, Jang SW, Ko J. Regulation of ADP-ribosylation factor 4 expression by small leucine zipper protein and involvement in breast cancer cell migration. Cancer Lett 2012; 314: 185-97.
44.
Alhammad R. Bioinformatics identification of TUBB as potential prognostic biomarker for worse prognosis in ER-positive and better prognosis in ER-negative breast cancer. Diagnostics (Basel) 2022; 12: 2067.
45.
Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol 1997; 13: 83-117.
46.
Knossow M, Campanacci V, Khodja LA, Gigant B. The mechanism of tubulin assembly into microtubules: insights from structural studies. iScience 2020; 23: 101511.
47.
El Amri M, Fitzgerald U, Schlosser G. MARCKS and MARCKS-like proteins in development and regeneration. J Biomed Sci 2018; 25: 43.
48.
Wang J, Jarrett J, Huang CC, Satcher RL Jr, Levenson AS. Identification of estrogen-responsive genes involved in breast cancer metastases to the bone. Clin Exp Metastasis 2007; 24: 411-22.
49.
Liang W, Gao R, Yang M, et al. MARCKSL1 promotes the proliferation, migration and invasion of lung adenocarcinoma cells. Oncol Lett 2020; 19: 2272-80.
50.
Karagoz K, Lehman HL, Stairs DB, Sinha R, Arga KY. Proteomic and metabolic signatures of esophageal squamous cell carcinoma. Curr Cancer Drug Targets 2016.
51.
Björkblom B, Padzik A, Mohammad H, et al. c-Jun N-terminal kinase phosphorylation of MARCKSL1 determines actin stability and migration in neurons and in cancer cells. Mol Cell Biol 2012; 32: 3513-26.
52.
Kosti A, de Araujo PR, Li WQ, et al. The RNA-binding protein SERBP1 functions as a novel oncogenic factor in glioblastoma by bridging cancer metabolism and epigenetic regulation. Genome Biol 2020; 21: 195.
53.
Guo K, Zheng S, Xu Y, Xu A, Chen B, Wen Y. Loss of miR-26a-5p promotes proliferation, migration, and invasion in prostate cancer through negatively regulating SERBP1. Tumour Biol 2016; 37: 12843-54.
54.
Wang T, Xu L, Jia R, Wei J. MiR-218 suppresses the metastasis and EMT of HCC cells via targeting SERBP1. Acta Biochim Biophys Sin 2017; 49: 383-91.
55.
Sunkara KP, Gupta G, Hansbro PM, Dua K, Bebawy M. Functional relevance of SATB1 in immune regulation and tumorigenesis. Biomed Pharmacother 2018; 104: 87-93.
56.
Panchal O, Wichmann G, Grenman R, et al. SATB1 as oncogenic driver and potential therapeutic target in head & neck squamous cell carcinoma (HNSCC). Sci Rep 2020; 10: 8615.
57.
Tesone AJ, Rutkowski MR, Brencicova E, et al. Satb1 overexpression drives tumor-promoting activities in cancer-associated dendritic cells. Cell Rep 2016; 14: 1774-86.
58.
Bied M, Ho WW, Ginhoux F, et al. Roles of macrophages in tumor development: a spatiotemporal perspective. Cell Mol Immunol 2023; 20: 983-92.
59.
Li H, Fan J, Ma J, Qiao L, Sha T, Ma B. Exosomal miR-182-5p from breast cancer cells reprogram tumor-associated macrophages and promote triple-negative breast cancer progression by targeting Notch1 in macrophages. Arch Med Sci 2024. doi:10.5114/aoms/187080.
60.
Zhang QW, Liu L, Gong CY, et al. Prognostic significance of tumor-associated macrophages in solid tumor: a meta-analysis of the literature. PLoS One 2012; 7: e50946.
61.
Zając AE, Czarnecka AM, Rutkowski P. The role of macrophages in sarcoma tumor microenvironment and treatment. Cancers 2023; 15: 5294.
62.
Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol 2000; 164: 6166-73.
63.
Ling H, Yang Z, Sun Y, et al. The impact of diffuse large B-cell lymphoma-derived exosomes on macrophage polarisation and cytokine release. Arch Med Sci 2020. doi:10.5114/aoms.2020.97355.
64.
Li M, He L, Zhu J, Zhang P, Liang S. Targeting tumor-associated macrophages for cancer treatment. Cell Biosci 2022; 12: 85.
65.
Mantovani A, Allavena P, Marchesi F, et al. Macrophages as tools and targets in cancer therapy. Nat Rev Drug Discov 2022; 21: 799-820.
66.
Almanza G, Searles S, Zanetti M. Delivery of miR-214 via extracellular vesicles downregulates Xbp1 expression and pro-inflammatory cytokine genes in macrophages. Extracell Vesicles Circ Nucl Acids 2024; 5: 249-58.
67.
Zhou L, Zhao T, Zhang R, Chen C, Li J. New insights into the role of macrophages in cancer immunotherapy. Front Immunol 2024; 15: 1381225.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.