Introduction
Colorectal cancer (CRC) is known as the most common gastrointestinal malignancy and one of the leading causes of cancer death worldwide [1, 2]. The survival of CRC patients has been improved by radical resection combined with adjuvant chemoradiotherapy [3], but recurrence or distant metastasis of CRC occurs in most patients and is the primary cause of treatment failure [4]. Therefore, a suitable indicator of the prognosis of CRC patients is pivotal to treatment in the clinic.
Translocase of the inner mitochondrial membrane 50 (TIMM50) is an essential component of the TIM23 complex, which mediates the translocation of transit peptide-containing proteins across the mitochondrial inner membrane [5, 6]. In humans, two isoforms of TIMM50 have been reported: a short TIMM50S (353 amino acids) isoform and a long TIMM50L (456 amino acids) isoform. TIMM50S exists in mitochondria as part of the TIM23 complex, whereas TIMM50L targets the nucleus due to the presence of an internal nuclear localization signal [7–9]. TIMM50 has been studied in tumour progression. Gao et al. [10] revealed that loss of TIMM50 suppressed cancer cell proliferation and induced cancer cell apoptosis in breast cancer. Zhang et al. [11] also found that overexpression of TIMM50 promoted tumour progression by phosphorylating ERK/P90RSK in non-small cell lung cancer cells, and they predicted TIMM50 to be a prognostic marker in non-small cell lung cancer patients. Nevertheless, whether TIMM50 is a biomarker for predicting the prognosis of CRC patients remains unknown.
To investigate the prognostic role of TIMM50 expression and assess the potential connection between TIMM50 expression and clinicopathological characteristics in CRC, we performed immunohistochemistry on 192 CRC patient samples; we found that TIMM50 may be a sensitive biomarker for the prognosis of CRC patients.
Material and methods
Patients and tissue samples
Colorectal cancer patients (123 men and 69 women) who had undergone radical resection from May 2013 to April 2018 participated in this study. None of the patients received chemoradiotherapy prior to surgery. The patients with colon cancers underwent the same surgical procedure for complete mesocolic excision. The patients with rectal cancers underwent the same surgical procedure for total mesorectal excision. This study was approved by the Ethics Committee of Huashan Hospital. Written consent was provided by all participants in this study. Tumour and normal tissues were procured from patients involved in this study. The tissues were snap-frozen after resection and stored in liquid nitrogen. The patients were followed up every 3 months after surgery for 5 years. During follow-up, regular evaluations included medical history, physical examination, complete blood count, thoracic computed tomography (CT), and abdominal ultrasound or CT. Disease progression was determined by the treating physician based on the available information, including clinical assessments, radiologic examination, and pathology reports.
Immunohistochemistry analysis of TIMM50
Immunohistochemistry (IHC) was performed strictly in accordance with the manufacturer’s instructions. In brief, tissue sections were cut to 3-µm thickness and placed on slides. The slides were incubated for 1 h at 72°C, immersed in xylene twice and rehydrated with graded ethanol. The slides were washed with PBS 5 times. The sections were immersed in sodium citrate buffer (pH 6.0) for 30 min at 95°C to enhance antigen retrieval. The sections were placed in 3% hydrogen peroxide for 10 min at room temperature to block endogenous peroxidase activity. Then, the slices were incubated for 24 h with a specific primary antibody for TIMM50 (1 : 100 dilution; Santa Cruz Biotechnology Inc., Santa Cruz, USA). After washing with PBST buffer, the slices were incubated with a goat anti-mouse biotinylated antibody for 1 h. Colour development was carried out using 3,3ʹ-diaminobenzidine (DAB), and counterstaining was performed using haematoxylin. After staining, the sections were dehydrated with increasing concentrations of ethanol and xylene.
The staining results were evaluated by two experienced pathologists in a double-blind manner. A semi-quantitative method was applied to judge the percentage of positive cells under the microscope and the staining intensity. The number of positive cells was observed in 5 high power fields (×200) for each slice, and the percentage of positive cells was counted. Positive colour intensity was scored as 0 (normal staining), 1 (weak staining), 2 (intermediate staining), and 3 (strong staining). The percentage of positive cells was scored as 1 (< 25%), 2 (26–50%), 3 (51–75%) and 4 (> 75%) [3]. The two scores were multiplied to obtain the final score. TIMM50 expression was defined as low for a score < 6 and high for a score > 6.
Quantitative real-time PCR
Total RNA was extracted from tumour tissues and adjacent tissues with Trizol homogenization buffer (Takara Bio. Inc., Takara, Japan); then, according to the PrimeScript RT reagent kit protocol (Takara Bio. Inc.), RNA was reverse transcribed into cDNA. The cDNA was amplified by quantitative real-time PCR using specific primers for TIMM50 (forward: 5ʹ-TTCCTGATGAGTTCGACAATG-3ʹ; reverse: 5ʹ-AGCTC-CAAAACGAGCGTGTA-3ʹ) and GAPDH (forward: 5’-CTGACTTCAACAGCGACACC-3ʹ; reverse: 5ʹ-TAG CCAAATTCGTTGTCATACC-3ʹ). The reaction volume was 20 µl (including 10 µl of SYBR green mix, 0.5 µl of each primer, 1 µl of cDNA, and 8 µl of ddH2O), and samples were detected with an ABI 7500 Real-Time PCR system. GAPDH was used as an internal reference gene, and the relative TIMM50 expression was determined using the Livak method (2–ΔΔCt).
Western blotting
The tumour and adjacent tissue samples were lysed completely in extraction buffer. The supernatants were centrifuged and collected. The protein concentrations were determined according to the BCA protocol. Equal amounts of protein samples (30 µg) were separated by 10% SDS-PAGE. Then, the proteins were transferred onto polyvinylidene fluoride (PVDF) membranes. Next, the PVDF membranes were incubated with primary antibodies at 4°C overnight, including TIMM50 and GAPDH (1 : 1000, Santa Cruz Biotechnology Inc., Santa Cruz, USA). Then, the membranes were incubated at room temperature with HRP-conjugated secondary antibodies (1 : 10,000 dilution) for 2 h. The results were obtained using an ECL substrate (Beyotime Biotechnology, Beyotime, China) detected by a ChemiScope 3300 Mini system (Clinx Science Instruments Co. Ltd., Clinx, Shanghai, China). GAPDH acted as a loading control.
Statistical analysis
All data analyses were conducted with the SPSS 19.0 software package. All qRT-PCR values are shown as the mean ± SD. The correlation between TIMM50 expression and clinicopathologic characteristics was analysed using the χ2 test. Kaplan-Meier curves were constructed to evaluate survival, and the log-rank test was applied to analyse the statistical significance of differences. Univariate and multivariate logistic regression analyses were used to assess TIMM50 expression and overall survival. Multivariate analysis using the Cox regression model was used to evaluate independent prognostic factors for overall survival. Other statistical analyses were performed with a two-tailed unpaired t-test, and p < 0.05 indicated statistical significance.
Results
Immunohistochemical staining characteristics
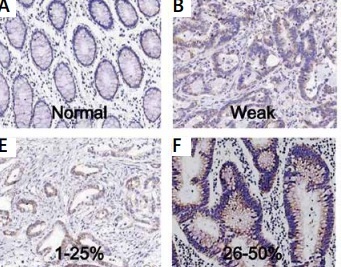
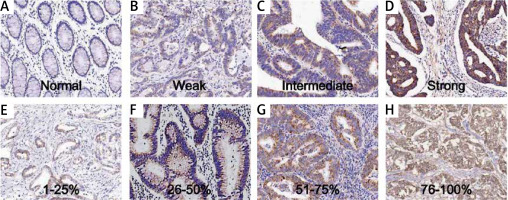
The samples were from 192 patients from our hospital. The expression of TIMM50 was detected by immunohistochemical staining. TIMM50 expression was assessed and scored. The TIMM50 immunohistochemical staining characteristics for colorectal cancers were as follows: staining intensity was 0 (Figure 1 A), 1 (Figure 1 B), 2 (Figure 1 C) and 3 (Figure 1 D). The percentage of positive cells was designated as 1 (1–25%) (Figure 1 E), 2 (26–50%) (Figure 1 F), 3 (51–75%) (Figure 1 G) and 4 (> 75%) (Figure 1 H).
TIMM50 expression was higher in colorectal cancer tissues than in paired adjacent normal tissues
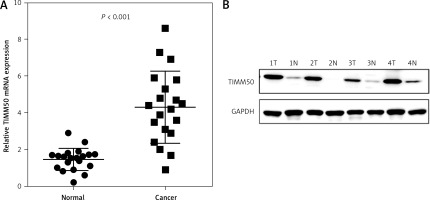
To investigate the potential value of TIMM50 as a prognostic biomarker in CRC, we determined the mRNA levels of TIMM50 by qRT-PCR in 20 samples from CRC patients, including 20 tumour tissues and paired adjacent normal tissues. We found that TIMM50 mRNA expression increased significantly in tumour tissues (Figure 2 A). Compared to that in paired adjacent normal tissues, increased protein expression of TIMM50 was identified in four tumour tissues (Figure 2 B), which was consistent with the TIMM50 mRNA expression results.
Correlation between TIMM50 expression and clinicopathologic variables
To further elucidate whether high TIMM50 expression affects the pathological development of CRC, we analysed the correlation between TIMM50 expression and clinicopathologic characteristics. As shown in Table I, high TIMM50 expression was significantly correlated with pathologic stage (p = 0.043), N stage (p = 0.048) and distant metastasis (p = 0.015), while TIMM50 expression was not related to other clinical factors, such as sex, age, tumour location, tumour size, T stage or preoperative serum CEA level.
Table I
Correlation between TIMM50 expression and clinicopathologic variables
| Characteristics | TIMM50 expression | P-value | ||
|---|---|---|---|---|
| All cases | Low | High | ||
| Age: | ||||
| ≥ 65 | 90 | 48 | 42 | 0.187 |
| < 65 | 102 | 64 | 38 | |
| Gender: | ||||
| Male | 123 | 70 | 53 | 0.593 |
| Female | 69 | 42 | 27 | |
| Tumor site: | ||||
| Colon | 92 | 53 | 39 | 0.845 |
| Rectum | 100 | 59 | 41 | |
| Grade: | ||||
| Well or moderate | 141 | 86 | 55 | 0.214 |
| Poor | 51 | 26 | 25 | |
| Pathologic stage: | ||||
| 1–2 | 91 | 60 | 31 | 0.043* |
| 3–4 | 101 | 52 | 49 | |
| T stage: | ||||
| 1–2 | 56 | 38 | 18 | 0.086 |
| 3–4 | 136 | 74 | 62 | |
| N stage: | ||||
| 0 | 93 | 61 | 32 | 0.048* |
| 1–2 | 99 | 51 | 48 | |
| Distant metastasis: | ||||
| 0 (no) | 167 | 103 | 64 | 0.015* |
| 1 (yes) | 25 | 9 | 16 | |
| CEA: | ||||
| ≤ 10 | 137 | 85 | 52 | 0.100 |
| > 10 | 55 | 27 | 28 | |
Prognostic value of TIMM50 in colorectal cancer
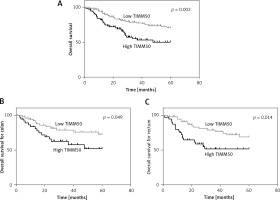
To understand the underlying value of TIMM50 as a prognostic biomarker in colorectal cancer, we analysed the expression levels of TIMM50 in tumour tissues from CRC patients. A Kaplan-Meier survival analysis indicated that patients who had low expression levels of TIMM50 had a longer overall survival than those who had high expression levels of TIMM50 (p = 0.002, Figure 3 A). According to different disease sites, we demonstrated that high TIMM50 expression in patients with colon cancer (p = 0.049, Figure 3 B) or rectal cancer (p = 0.014, Figure 3 C) resulted in a shorter overall survival according to a stratified analysis. Furthermore, we found that lymph node invasion (p = 0.020), distant metastasis (p = 0.001), high TNM stage (p = 0.025) and high TIMM50 expression (p = 0.002) were significantly related to overall survival according to a univariate analysis (Table II). Distant metastasis and high TIMM50 expression were confirmed as independent prognostic factors for the overall survival of CRC patients in a multivariate analysis using the Cox regression model (p = 0.003) (hazard ratio = 2.2, 95% CI: 1.3–3.7, p = 0.003) (Table III).
Table II
Assessment of the effect of TIMM50 expression in colorectal cancer and clinical manifestations on overall survival by univariate analysis model
| Variable | Univariate analysis | |
|---|---|---|
| HR (95% CI) | P-value | |
| Age (≥ 65) | 1.4 (0.8–2.2) | 0.244 |
| Tumor site (rectum) | 1.1 (0.7–1.9) | 0.667 |
| Poor differentiation | 1.7 (1.0–2.9) | 0.057 |
| T staging | 1.3 (0.7–2.3) | 0.349 |
| Lymph node invasion | 1.9 (1.1–3.1) | 0.020* |
| Distant metastasis | 2.8 (1.5–5.2) | 0.001* |
| Higher TNM staging | 1.8 (1.1–3.0) | 0.025* |
| Higher CEA level | 1.4 (0.8–2.4) | 0.188 |
| Higher TIMM50 expression | 2.2 (1.3–3.7) | 0.002* |
Table III
Assessment of the effect of TIMM50 expression in colorectal cancer and clinical manifestations on overall survival by multivariate analysis model
| Variables | Multivariate analysis | |
|---|---|---|
| HR (95% CI) | P-value | |
| Distant metastasis | 2.7 (1.5–5.1) | 0.001* |
| Higher TIMM50 expression | 2.2 (1.3–3.7) | 0.003* |
Figure 3
Colorectal cancer patients with high TIMM50 expression had poor overall survival. A Kaplan-Meier survival analysis indicated that patients who had low expression levels of TIMM50 had longer overall survival than those who had high expression levels of TIMM50 (A). According to different disease sites, patients with high TIMM50 expression and colon cancer (B) and rectal cancer (C) had significantly shorter overall survival according to stratified analysis
Discussion
In eukaryotes, mitochondria play a central role in oxidative metabolism, and a number of research studies have indicated that mitochondria also play an integral role in the cascade of intracellular events that lead to cell proliferation, programmed cell death and apoptosis, which may promote the occurrence and development of human malignancies [12–14]. The function of mitochondria depends on the interaction between mitochondria and nuclei. In total, 99% of the thousands of mitochondrial proteins are encoded by nuclear genes, which are synthesized in the cytoplasm and transported to mitochondrial components through the mitochondrial membrane protein transport system [15, 16]. A variety of mitochondrial DNA replication factors, transcriptional regulators, mitochondrial DNA mutation repair enzymes, and dismutases that cleave mitochondrial oxygen free radicals are encoded by nuclear genes, and proper mitochondria entry is a prerequisite for their important roles [17, 18].
It has been found that abnormalities in the mechanism of mitochondrial membrane protein transport processing are associated with the development of various diseases [14, 16, 19]. Mitochondrial encephalopathies in most reported patients with isolated complex V deficiency are due to mutations in TMEM70, which is related to mitochondrial transport proteins [20, 21]. Mendonca et al. [22] revealed that mitochondrial transport protein TSPO expression was more obvious as the degree of malignancy increased, and this expression was negatively correlated with the prognosis and survival time of glioma patients. Compared with that in initial glioma, TSPO expression was increased and associated with increased malignancy. Inherited mitochondrial DNA mutations were involved in CRC [23]. However, the mechanism of mitochondrial membrane protein transport in CRC is unclear. In our study, we focused on translocase of the inner mitochondrial membrane, TIMM50. The TIM23 complex is composed of at least TIMM23, TIMM17 (TIMM17A or TIMM17B) and TIMM50. This complex interacts with the TIMM44 component of the PAM complex, and with DNAJC15, TIMM50 plays an important role in mitochondrial membrane proteins [24]. We found that mRNA and protein expression levels of translocase of the inner mitochondrial membrane, TIMM50, were significantly increased in CRC tissues compared to those in paired adjacent normal tissues, which was in accordance with the IHC results. Moreover, a correlation analysis suggested that high TIMM50 expression was significantly associated with pathologic stage, N stage and distant metastasis, while TIMM50 expression was not related to other clinical factors.
Over the past two decades, the search for important CRC-related biomarkers has attracted attention, and several important prognostic biomarkers have been described [25–28]. Andreyev et al. [29] showed that Kirsten ras was associated with a poor prognosis in 2445 CRC patients and that Kirsten ras mutations were associated with an increased risk of relapse and death. A multivariate analysis suggested that the presence of a mutation increased the risk of recurrence. Richman et al. [30] stated that mutations in KRAS or BRAF in advanced CRC were a poor prognostic factor for overall survival but had a minimal impact on progression-free survival. All significant factors according to the univariate analysis were subjected to multivariate analysis, but only BRAF mutations remained an independent prognostic factor for reduced overall survival. Mei et al. [3] revealed that high expression of interferon regulatory factor 2 (IRF-2) was correlated with significantly worse overall survival and that IRF-2 was a prognostic biomarker in CRC. Therefore, it was necessary to refine prognostic information and improve the precision with which we are able to define the patient cohorts that will benefit from systemic treatment. According to our research results, a Kaplan-Meier survival analysis indicated that patients with high expression of TIMM50 had poorer malignant progression of CRC than those with low expression of TIMM50. Additionally, a multivariate analysis suggested that distant metastasis and higher TIMM50 expression were independent prognostic factors for overall survival. Some patients received postoperative chemoradiotherapy according to NCCN guidelines. Different postoperative treatment options also affect overall survival of CRC patients. Since our sample size is not very large, we did not perform further subgroup analysis. The current results highlight that TIMM50 could be a prognostic factor for CRC, which could be a new indicator for the prognosis of CRC patients.
According to previous reports, TIMM50 participates in regulating the tumour invasion and proliferation of cancer [10]. Zhang et al. [11] demonstrated that TIMM50 promoted tumour invasion and proliferation in non-small cell lung cancer cells by upregulating cyclin D1 and downregulating E-cadherin. The researchers also found that TIMM50 was able to upregulate ERK and P90RSK phosphorylation. The molecular mechanism of TIMM50 as a prognostic factor in CRC requires further research.
In conclusion, TIMM50 may be a key factor for monitoring CRC and a new indicator for CRC patient prognosis. In addition, these results imply that a potential therapeutic target that regulates the expression level of TIMM50 could decrease the malignant progression of CRC and improve the overall survival of CRC patients.