Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Section Editors
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
CLINICAL RESEARCH
Novel biotargets of colorectal cancer peritoneal metastasis
1
Department of Gastroenterology, Affiliated Hangzhou First People’s Hospital, Westlake University School of Medicine, Hangzhou, Zhejiang, China
2
Hangzhou Hospital and Institute of Digestive Diseases, Hangzhou, Zhejiang, China
3
Key Laboratory of Integrated Traditional Chinese and Western Medicine for Biliary and Pancreatic Diseases of Zhejiang Province, Hangzhou, Zhejiang, China
4
Department of Radiotherapy, BenQ Medical Center, The Affiliated BenQ Hospital of Nanjing Medical University, Nanjing, Jiangsu Province, China
5
Beijing Etown Academy, Beijing, China
Submission date: 2023-08-16
Final revision date: 2023-09-22
Acceptance date: 2023-10-17
Online publication date: 2024-12-20
Corresponding author
Xiaofeng Zhang
Department of Gastroenterology, Affiliated Hangzhou First People’s Hospital, Westlake University School of Medicine Hangzhou, Zhejiang 310006, China, Phone: +86-0571-56006782
Department of Gastroenterology, Affiliated Hangzhou First People’s Hospital, Westlake University School of Medicine Hangzhou, Zhejiang 310006, China, Phone: +86-0571-56006782
KEYWORDS
TOPICS
ABSTRACT
Introduction:
Peritoneal metastasis often predicts advanced progression and a poor prognosis in colorectal cancer (CRC). However, peritoneal metastases are extremely difficult to predict or diagnose by routine diagnostic methods.
Material and methods:
In this study, a microarray containing 30 samples from peritoneal metastasis and their matched CRC primaries obtained during cytoreductive surgery were compared to take a long hard look at all the options on the significant differentially expressed genes. The potential interactions and mechanisms of these expressed genes in promoting peritoneal metastasis were analyzed and studied by multiple bioinformatics analysis.
Results:
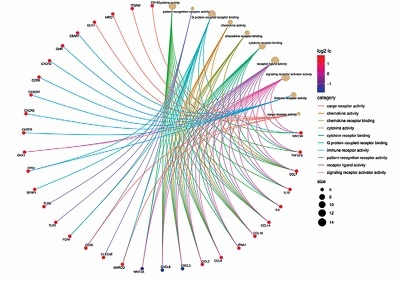
The results suggested that the functions of these genes are closely related to immune response and cytokine activity. Additionally, the top 10 core genes’ correction with the leukocyte infiltration and serum cytokine profiles were identified and may be expected to become diagnostic and therapeutic targets of peritoneal metastasis in CRC.
Conclusions:
The expression of IL-6, IL-10 and IL-17 in plasma and their correlation with leukocyte infiltration are proven potential diagnostic, prognostic, and therapeutic biomarkers for peritoneal management of CRC.
Peritoneal metastasis often predicts advanced progression and a poor prognosis in colorectal cancer (CRC). However, peritoneal metastases are extremely difficult to predict or diagnose by routine diagnostic methods.
Material and methods:
In this study, a microarray containing 30 samples from peritoneal metastasis and their matched CRC primaries obtained during cytoreductive surgery were compared to take a long hard look at all the options on the significant differentially expressed genes. The potential interactions and mechanisms of these expressed genes in promoting peritoneal metastasis were analyzed and studied by multiple bioinformatics analysis.
Results:
The results suggested that the functions of these genes are closely related to immune response and cytokine activity. Additionally, the top 10 core genes’ correction with the leukocyte infiltration and serum cytokine profiles were identified and may be expected to become diagnostic and therapeutic targets of peritoneal metastasis in CRC.
Conclusions:
The expression of IL-6, IL-10 and IL-17 in plasma and their correlation with leukocyte infiltration are proven potential diagnostic, prognostic, and therapeutic biomarkers for peritoneal management of CRC.
REFERENCES (37)
1.
Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin 2020; 70: 145-64.
2.
Miller KD, Nogueira L, Devasia T, et al. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin 2022; 72: 409-36.
3.
Segelman J, Granath F, Holm T, et al. Incidence, prevalence and risk factors for peritoneal carcinomatosis from colorectal cancer. J Br Surg 2012; 99: 699-705.
4.
Franko J, Shi Q, Meyers JP, et al. Prognosis of patients with peritoneal metastatic colorectal cancer given systemic therapy: an analysis of individual patient data from prospective randomised trials from the Analysis and Research in Cancers of the Digestive System (ARCAD) database. Lancet Oncol 2016; 17: 1709-19.
5.
Vassos N, Piso P. Metastatic colorectal cancer to the peritoneum: current treatment options. Curr Treat Options Oncol 2018; 19: 49.
6.
Koh JL, Yan TD, Glenn D, et al. Evaluation of preoperative computed tomography in estimating peritoneal cancer index in colorectal peritoneal carcinomatosis. Ann Surg Oncol 2009; 16: 327-33.
7.
Lenos KJ, Bach S, Ferreira Moreno L, et al. Molecular characterization of colorectal cancer related peritoneal metastatic disease. Nat Commun 2022; 13: 4443.
8.
Wu D, Rice CM, Wang X. Cancer bioinformatics: a new approach to systems clinical medicine. BMC Bioinformatics 2012; 13: 71.
10.
Kadoch C, Hargreaves DC, Hodges C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 2013; 45: 592-601.
11.
Patra BG, Maroufy V, Soltanalizadeh B, et al. A content-based literature recommendation system for datasets to improve data reusability – a case study on Gene Expression Omnibus (GEO) datasets. J Biomed Inform 2020; 104: 103399.
12.
Barriuso J, Nagaraju RT, Belgamwar S, et al. Early adaptation of colorectal cancer cells to the peritoneal cavity is associated with activation of ‘stemness’ programs and local inflammation. Am Assoc Cancer Res 2021; 27: 1119-30.
13.
Huang DW, Sherman BT, Tan Q, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 2007; 8: R183.
14.
Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019; 10: 1523.
15.
Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010; 38 (Suppl_2): W214-20.
16.
Saito R, Smoot ME, Ono K, et al. A travel guide to Cytoscape plugins. Nat Methods 2012; 9: 1069-76.
17.
Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 2010; 39 (Suppl 1): D561-8.
18.
Cline MS, Smoot M, Cerami E, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2007; 2: 2366-82.
19.
Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014; 8 Suppl 4: S11.
20.
Li T, Fan J, Wang B, et al. TIMER: A Web Server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 2017; 77: e108.
21.
Klaver Y, Simkens L, Lemmens V, et al. Outcomes of colorectal cancer patients with peritoneal carcinomatosis treated with chemotherapy with and without targeted therapy]. Eur J Surg Oncol 2012; 38: 617-23.
22.
Klaver YLB, Lemmens VEPP, De Hingh IHJT. Outcome of surgery for colorectal cancer in the presence of peritoneal carcinomatosis. Eur J Surg Oncol 2013; 39: 734-41.
23.
Chidlow JH, Glawe JD, Alexander JS, et al. VEGF164 differentially regulates neutrophil and T cell adhesion through ItgaL- and ItgaM-dependent mechanisms. Am J Physiol Gastrointest Liver Physiol 2010; 299: G1361-7.
24.
Saraiva M, O’garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol 2010; 10: 170-81.
26.
Gabay C. Interleukin-6 and chronic inflammation. Arthritis Res Ther 2006; 8 (2 Suppl): S3.
27.
Wang S, Sun Y. The IL-6/JAK/STAT3 pathway: potential therapeutic strategies in treating colorectal cancer. Int J Oncol 2014; 44: 1032-40.
28.
Lu S, Pardini B, Cheng B, et al. Single nucleotide polymorphisms within interferon signaling pathway genes are associated with colorectal cancer susceptibility and survival. PLoS One 2014; 9: e111061.
29.
Linsley PS, Ledbetter JA. The role of the CD28 receptor during T cell responses to antigen. Ann Rev Immunol 1993; 11: 191-212.
30.
Lenschow DJ, Walunas TL, Bluestone JA. CD28/B7 system of T cell costimulation. Ann Rev Immunol 1996; 14: 233-58.
31.
Slavik JM, Hutchcroft JE, Bierer BE. CD28/CTLA-4 and CD80/CD86 families: signaling and function. Immunol Res 1999; 19: 1-24.
32.
Chowdhury D, Lieberman J. Death by a thousand cuts: granzyme pathways of programmed cell death. Annu Rev Immunol 2008; 26: 389-420.
33.
Schön M, Schön M. TLR7 and TLR8 as targets in cancer therapy. Oncogene 2008; 27: 190-9.
34.
Diebold SS, Kaisho T, Hemmi H, et al. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 2004; 303: 1529-31.
35.
Figdor CG, Van Kooyk Y, Adema GJ. C-type lectin receptors on dendritic cells and Langerhans cells. Nat Rev. Immunol 2002; 2: 77-84.
36.
Minter LM, Turley DM, Das P, et al. Inhibitors of -secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nat Immunol 2005; 6: 680-8.
37.
Shen Y, Nussbaum YI, Manjunath Y, et al. TBX21 methylation as a potential regulator of immune suppression in CMS1 subtype colorectal cancer. Cancers 2022; 14: 4594.
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.