Introduction
Atherosclerosis, a chronic progressive disease, is characterized by fibrous and lipid plaques that narrow and harden arteries, thus resulting in various diseases such as coronary heart disease, peripheral artery disease and stroke. Two main factors, lipids and inflammation, are considered to be involved in the formation of atheromas [1–3]. In the past, the main strategies for treating atherosclerosis focused on regulating lipid metabolism and lowering low-density lipoprotein. Recently, some anti-inflammatory drugs began to be used to treat atherosclerosis [4–6]. Nevertheless, the choice of anti-inflammatory drugs is very limited. For instance, some non-steroidal anti-inflammatory drugs may even increase the risk of cardiovascular events [4]. Therefore, finding new anti-atherosclerotic drugs and determining their mechanisms are crucial.
Artemisinin (ART), which was first extracted from Artemisia annua by Tu Youyou, has been used as an efficient drug for curing malaria [7]. In addition to its anti-malarial activity, ART can be used to treat other diseases such as neuroinflammation, cancer and lupus erythematosus, owing to its outstanding anti-inflammatory and anti-angiogenetic characteristics [8, 9]. Hence, we hypothesized that ART might alleviate atherosclerosis. We were excited to find that ART inhibited the proliferation of vascular smooth muscle cells (VSMCs) in vitro and alleviated atherosclerotic plaque formation in vivo [10–12]. Nonetheless, ART is not clinically used to treat atherosclerosis because the mechanism of ART against atherosclerosis was unclear.
Long non-coding RNAs (lncRNAs) are linear RNAs that do not encode proteins and used to be considered junk RNA. In recent years, researchers have found that numerous lncRNAs are involved in the regulation of the cell cycle, proliferation and apoptosis [13, 14]. Herein, we hypothesized that lncRNAs are engaged in the effects of ART on atherosclerosis. Through high-throughput sequencing techniques, we detected the differentially expressed lncRNA profiles in an atherosclerotic model mice treated with ART (ART group) and atherosclerosis model mice without ART treatment (AS group), and explored the possible mechanism through which ART alleviates atherosclerosis in mammals. This study aims to provide a theoretical basis for the future use of artemisinin in the treatment of atherosclerosis.
Material and methods
Ethical statement
This study was approved by the Animal Ethics Committee of Shengjing Hospital (Ethic No. 2018PS513K). Animal experiments followed the guidelines of the European Convention for the Protection of Vertebrate Animals used for Experimental and Other Scientific Purposes and the Animal Research: Reporting of In Vivo Experiments guidelines.
Groups, atherosclerosis modeling and ART treatment
Eight-week-old male C57BL6 mice and apolipoprotein E deficient (APOE–/–) mice with the same genetic background were purchased from Beijing Huafukang Biotechnology Co., Ltd. (Beijing, PR China). Eight C57BL/6 mice fed a standard rodent chow diet served as the control group. Sixteen APOE–/– mice were randomly divided into two groups, eight in the atherosclerosis group and eight in the ART group. Mice in the atherosclerosis group were given a Western diet (21% fat, 0.15 cholesterol, Beijing Huafukang Biotechnology, No. H10141). Mice in the ART group were given a Western diet and were administered ART (purity > 98%, Beijing Solarbio Science & Technology Co., Ltd., IA0520) orally at 50 mg/kg/day as described previously [15]. The three groups were housed in a temperate room at a temperature of 22 ±1°C and humidity of 45–55%. The treatment process described above lasted 8 weeks.
Specimen acquisition
Specimen acquisition was performed when the mice were 16 weeks old. All mice were euthanized through intraperitoneal injection of pentobarbital. The hearts of the mice were separated and fixed in 4% paraformaldehyde for Oil Red O staining. The intact aortas of the mice of the atherosclerosis group and ART group were carefully separated from the surrounding adherent tissue and stored in liquid nitrogen for the bioinformatics analysis.
Oil Red O staining
To visualize the atherosclerotic lipid accumulation, we stained the heart slices of the mice with Oil Red O (Sigma-Aldrich O-0625). In brief, the heart was cut into sections perpendicular to the long axis of the heart, and the aortic valve plane was chosen to perform Oil Red O staining. After rinsing with phosphate buffered saline, the aortic valve slices were stained with Oil Red O for 25 min at room temperature. Then the slices were rinsed with 60% isopropyl alcohol and distilled water. Ultimately, the images of Oil Red O staining aortic valve slices were obtained with a microscope (Nikon Eclipse NI, Japan). Positively stained areas of five sections from each mouse were quantified through ImageJ by a researcher who was blind to the experimental condition and the results were presented as the mean lesion area.
RNA extraction and quality control
Total RNA was extracted from the aortas of the mice with RNAiso Plus (Takara Bio Inc., Japan) in accordance with the protocol. The quality and quantity of RNA were assessed with a NanoDrop ND-1000 spectrophotometer. The OD260/280 ratios of all samples were between 1.8 and 2.
High-throughput sequencing
The lncRNA and mRNA libraries were generated from the total RNAs extracted from aorta samples of three randomly selected mice in the atherosclerosis group and ART group with an rRNA-depleted Preparation Kit for Illumina (Neb, USA), according to the protocol. Then the libraries were sequenced on the Illumina HiSeq 4000 platform according to the vendor’s instructions by NovoGene Genome Sequencing Company (Beijing, China).
The ceRNA network construction
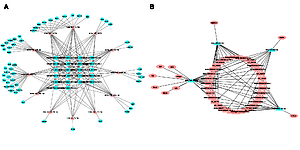
LncRNA-miRNA networks were constructed in StarBase v3.0. Then we used StarBase v3.0 and TargetScan v7.2 to predict the miRNA-mRNA interactions. LncRNA-miRNA-mRNA networks conforming to the competing endogenous RNAs (ceRNA) rules were depicted in Cytoscape v3.7.1.
KEGG and GO analysis
Gene Ontology (http://www.geneontology.org) analysis of biological processes, cellular components and molecular functions was used to classify the functions of differentially expressed mRNAs. Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg) pathway analysis was used to indicate the main biochemical and signaling pathways in which differentially expressed genes participate.
Quantitative real-time polymerase chain reaction
We selected RNA samples from 5 mice per group for validation. cDNA was synthesized by using the PrimeScript RT with gDNA Eraser reagent kit (Takara, RR047A). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed with 2 µl of cDNA, TB Green Premix Ex Taq II (Takara, RR820A), and gene specific primers which were synthesized by Sangon Biotech (Shanghai, China) (Table I). β-actin was used as a reference. The specificity was confirmed on the basis of the single peak on the melting curve. Three individual technical replicates were examined for the sample. The relative levels normalized to β-actin were calculated with the 2–ΔΔCT method.
Table I
Primers of lncRNAs
Results
ART alleviated atherosclerosis in APOE–/– mice
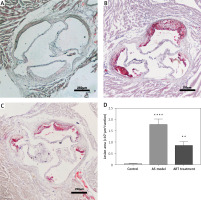
We established three groups: a control group consisting of C57BL/6 mice, an AS group consisting of high-fat diet fed APOE–/– mice without ART treatment and an ART group consisting of high-fat diet fed APOE–/– mice with ART treatment. Oil red O staining was used to evaluate the severity of aortic atherosclerosis. The area of atherosclerotic plaques in the atherosclerosis group (Figure 1 B; 1.77 ±0.14 mm²) was significantly larger than that in the control group (Figure 1 A; 0.04 ±0.02 mm²; p < 0.0001), thus confirming the success of atherosclerosis modeling (Figure 1 D). Compared with that in the atherosclerosis group, there was a significantly smaller area of atherosclerotic plaques in the ART group (Figure 1 C; 0.83 ±0.10 mm²; p < 0.01). This result illustrated the anti-atherosclerosis effect of ART and laid a foundation for subsequent bioinformatics analyses.
Figure 1
Artemisinin alleviated atherosclerosis. The area of atherosclerotic plaques in the atherosclerosis group (B) was significantly larger (p < 0.0001) than that in the control group (A). The area of plaques in the ART group (C) was significantly smaller (p < 0.01) than that in the atherosclerosis group. D – Statistical results
**p < 0.001.
Screening of differentially expressed lncRNAs and mRNAs
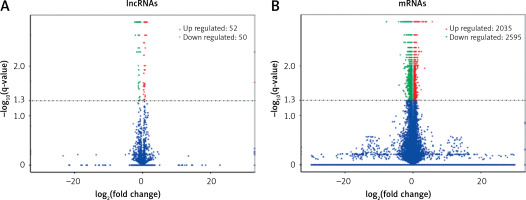
The expression profiles were compared between the main artery samples of the ART group and atherosclerosis group. Among a total of 3625 lncRNAs and 55831 mRNAs detected by high-throughput sequencing, there were significant differences in the expression of 102 lncRNAs and 4630 mRNAs (p < 0.05). Furthermore, 22 lncRNAs decreased and 5 lncRNAs increased in the ART group by showing significantly differential expression with an absolute log2 (fold change) > 1 (p < 0.05) (Table II). Among these differentially expressed lncRNAs, 50 lncRNAs decreased, and 52 lncRNAs increased in the ART group (Figure 2 A). Moreover, compared with the atherosclerosis group, in the ART group, 2595 mRNAs decreased, and 2035 mRNAs increased (Figure 2 B). The top 10 differentially expressed mRNAs are listed in Table III.
Table II
Differentially expressed lncRNA
Table III
Top 10 mRNAs
Predicted networks of ceRNAs
The networks of ceRNAs were constructed on the basis of increased and decreased lncRNAs. By using StarBase v3.0, among those differentially expressed lncRNAs, 35 upregulated and 22 downregulated lncRNAs can be predicted to have their potential binding miRNAs. Then the predicted miRNAs whose trends were compatible with the high-throughput sequencing data were input into two databases (StarBase v3.0 and TargetScan v7.2) to predict the interaction of miRNAs and mRNAs. In the same way, the predicted mRNAs whose trends were compatible with the high-throughput sequencing data were filtered. Ultimately, with Cytoscape v3.7.1, we determined and depicted the relevance of lncRNAs, miRNAs and mRNAs (Figure 3). Twelve increased miRNAs and four decreased miRNAs in the ART group were found in the ceRNA network. LncRNA ENSMUST00000099676.4 was predicted to interact with miR-370. LncRNA ENSMUST00000143673.1 and lncRNA ENSMUST00000070085.5 were predicted to interact with let-7a. LncRNA ENSMUST00000224554 was predicted to interact with miR-145. The predicted downstream miRNAs mentioned above have been reported to be associated with AS.
GO and KEGG analysis
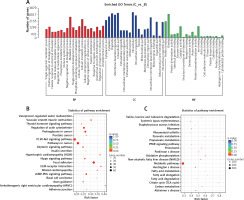
GO analysis indicated that specific mRNAs involved in biological processes (e.g., metabolic process and biological regulation), cellular components (e.g., organelle and intracellular) and molecular functions (e.g., binding and catalytic activity) were enriched. The KEGG analysis illustrated that these differentially expressed mRNAs were associated with atherosclerosis, through categories including amino acid degradation, oxidative phosphorylation, peroxisome, fatty acid metabolism and the cGMP-PKG signaling pathway (Figure 4).
Figure 4
Go and KEGG analysis. A – GO analysis of differentially expressed mRNAs. B – KEGG pathway analysis of increased mRNAs in ART group. C – KEGG analysis of decreased mRNAs in ART group. The enrichment factor, which is the ratio of the number of differentially expressed genes in the pathway and the number of all genes annotated in the pathway, indicates the reliability of the enrichment. Q-value, which is a normalized p-value, indicates the significance of the enrichment
Validation of differentially expressed lncRNAs
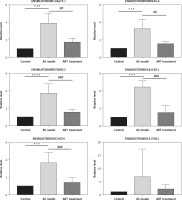
To validate the high-throughput sequencing results, we used qRT-PCR to detect six decreased lncRNAs of interest. Compared with those in the atherosclerosis group, levels of five of six lncRNAs (ENSMUST00000099676.4, ENSMUST00000143673.1, ENSMUST00000070085.5, ENSMUST00000182520.1, and ENSMUST00000-224554) were lower in the ART group. However, the expression level of lncRNA ENSMUST-00000141700.1 showed no significant difference between the ART group and atherosclerosis group (Figure 5).
Discussion
Atherosclerosis imposes a worldwide medical disease burden, especially in developed countries [16]. Although anti-lipid drugs and anti-inflammatory drugs are widely used to cope with this severe medical burden, the presence of numerous side effects makes those drugs not suitable for all people. Thus, finding a new treatment with few side effects has become a focus of attention. Recently, ART, an excellent anti-malaria drug with high safety and low side effects in clinic use, has been found to possess anti-atherosclerotic properties. However, the precise mechanism through which ART mitigates atherosclerosis remains unclear. More laboratory evidence is needed to confirm the anti-atherosclerotic capacity of ART, to lay a foundation for future clinical applications. Hence, in this study, by using transcriptome sequencing technology, we analyzed the possible mechanism through which ART alleviates atherosclerosis in mammals.
Over the past decade, lncRNAs have been found to be involved in the regulation of critical physiological and pathological processes in cells via diverse mechanisms [13]. Recent studies have indicated that lncRNAs participate in atherosclerosis [14]. The lncRNA H19 is upregulated in the plasma of patients with atherosclerosis and regulates ox-LDL-induced endothelial cell injury by acting as a let-7a sponge [17]. Interestingly, in our study, lncRNA H19 was decreased, and let-7a was increased in the ART treatment group, thus indicating one possible treatment mechanism through which ART decreases atherosclerosis.
CeRNAs are a group of RNAs that regulate transcripts of other RNAs by competing for shared miRNAs. LncRNAs regulate the expression of target genes by serving as miRNA sponges via ceRNA networks. Thus, we predicted the ceRNA networks of decreased and increased lncRNAs. Most of the increased miRNAs predicted by the downregulated lncRNAs in the ART group have been reported to have a protective effect on inflammation and atherosclerosis. MiR-370, which increased in the ART treatment group and might function downstream of lncRNA ENSMUST00000099676.4, has been reported to inhibit vascular inflammation [18]. The predicted downstream mRNAs regulated by miR-370 have been reported to promote the formation of atherosclerosis. For instance, nucleotide-binding oligomerization domain-containing protein 1 (Nod1) accelerates atherogenesis [19]. Deletion of BH3-interacting domain death agonist (Bid) inhibits cell proliferation and protects endothelial cells from oxidative stress related injury [20]. Interferon regulatory factor 1 (IRF1) induces VCAM-1 expression and inflammatory reactions through the activation of inducible NO synthase [21]. MiR-3473 suppressed inflammation and macrophage activation by inducing expression of IL-10 [22]. The miR-152 decreased the proliferation of HUVECs and malignant atherosclerotic progression via downregulation of Kruppel-like factor 5 (KLF5) [23]. MiR-148b, regulated by lncRNA H19, suppressed the proliferation of oxLDL-stimulated VSMCs and induced apoptosis of VSMCs [24]. Both miR-145 and miR-143 alleviated the proliferation and migration of VSMCs [25]. Let-7a attenuated ox-LDL induced endothelium injury by inhibiting NF-κB nuclear translocation [26]. Thus, we hypothesized that ART might downregulate the formation of atherosclerosis through a ceRNA mechanism, on the basis of the decreased lncRNA in the sequencing results and the miRNAs mentioned above. Additionally, the decreased miRNAs, such as miR-150 and miR-134, predicted by the lncRNAs upregulated in the sequencing results, have been reported to enhance inflammatory reactions or atherosclerotic progression. Knockdown of miR-150 alleviated inflammatory damage by up-regulating KLF2 [27]. Prior studies have shown that miR-134, which has been reported to increase in atherosclerotic macrophages, activates lipid accumulation and the inflammatory response by regulating angiopoietin-like 4 (Angptl4) [28]. The miR-182 increased LPL expression and activates lipid accumulation in APOE–/– mice by regulating HDAC9 [29]. SH3 domain-binding protein 4 (SH3BP4), which is the predicted downstream mRNA of miR-182, was a negative regulator of amino acid-dependent mTORC1 signaling, thereby inhibiting cell growth and promoting autophagy [30]. The mRNAs and miRNAs mentioned above, which may be regulated by the lncRNAs obtained from the high-throughput sequencing results, are potential targets of ART for the treatment of atherosclerosis.
Aberrantly expressed mRNAs may reflect the functions of lncRNAs whose expression is altered by ART treatment. Thus, we conducted GO and KEGG analyses of differentially expressed mRNAs to reveal the functions of differentially expressed lncRNAs in ART treatment of atherosclerosis. GO analysis showed that differentially expressed mRNAs were linked to the functions of metabolic and immune system processes and protein binding associated with atherosclerosis, thus indicating that ART may relieve atherosclerosis by regulating these processes. The expression level of hemolytic complement (Hc), also known as C5a, which participates in metabolic processes and neovascularization, was downregulated in the ART treatment group [31]. Our study provides a basis for understanding the mechanism through which ART treatment mitigates atherosclerosis. KEGG analysis indicated that differentially expressed mRNAs were associated with several specific signaling pathways, such as amino acid degradation, metabolic processes, vascular smooth muscle contraction and the PI3K-Akt signaling pathway. Specific amino acids engage in the formation of macrophage foam cells. Leucine has been confirmed to have anti-atherogenic properties in vivo [32]. Downregulated mRNAs involved in degradation of valine, leucine and isoleucine in the ART-treated group may indicate elevated levels of leucine. Although leucine levels were not measured for technical reasons, KEGG analysis suggested a possible target for ART in the treatment of atherosclerosis. In our sequencing data, LNC_000243 (chr 11: 58183820–58187387) was co-localized with 4930438A08Rik, which is associated with specific amino acid degradation. We suggest that ART may alleviate atherosclerosis by regulating the LNC_000243/4930438A08Rik/leucine axis. Upregulated mRNA enrichment in vascular smooth muscle contraction and the PI3K-Akt signaling pathway indicated that the ability of smooth muscle cell proliferation and migration decreased in the ART treatment group. The above enrichment analysis may potentially support future research on the mechanism through which ART decreases atherosclerosis.
In conclusion, our sequencing results revealed that the expression of specific mRNAs, miRNAs and lncRNAs associated with atherosclerosis was altered in the ART treatment group. The relative expression levels of lncRNAs were validated through qRT-PCR. An lncRNA-miRNA-mRNA network was constructed to explore ceRNAs involved in ART treatment of atherosclerosis. GO and KEGG analyses were used to search for possible mechanisms of ART in the treatment of atherosclerosis. Our findings suggested a possible mechanism of ART in the treatment of atherosclerosis and provided clues regarding the potential roles of lncRNAs in ART treatment. In addition, this research provides a new basis and hypothesis regarding the clinical application of ART in the treatment of atherosclerosis. There were some limitations in this study. First, we did not conduct experiments to explore the function of those RNAs and the underlying mechanism. Second, the sample size was not large enough. Further detailed research is expected to clarify the mechanism through which ART treatment decreases atherosclerosis and to identify lncRNAs involved in ART treatment, thus providing a stronger theoretical basis for the future application of ART in patients with atherosclerosis.