Introduction
Microsatellites (MSs) are repeating DNA motifs, or short tandem repeats, closely associated with vital genes in the genome [1]. MSs are prevalent in non-coding gene regions and are implicated in chromosomal rearrangements that impact gene replication/expression. Due to their repetitive structures, MSs are prone to replication errors during DNA replication as the DNA polymerase enzyme may slip on the MS regions, eventually causing frame-shift mutations or protein truncations [1, 2]. Under normal conditions, mutations containing MS regions in genes such as TGFβ-R2, ILGF, E2F4, and BAX could be repaired by the mismatch repair (MMR) system. However, in tumor cells, the absence of MMR genes or the failure to synthesize seven related proteins (h-MLH1, h-MLH3, h-MSH2, h-MSH3, h-MSH6, h-PMS1, and h-PMS2), or errors in the replication repair process, can result in DNA-MMR deficiency and microsatellite instability (MSI) formation [1–3].
Tumors with MSI in over 30–40% of examined loci are classified as high MSI (MSI-H), those with MSI in less than 30–40% are low MSI (MSI-L), and those with no detectable MSI are microsatellite stable (MSS) [4]. MSI significantly influences the development and outcomes of various cancers, with MSI-H patients generally showing better anti-tumor effects, greater ability to inhibit tumor growth, and improved prognosis compared to MSI-L or MSS cases. MSI serves as a proven prognostic and recurrence indicator in colorectal cancer (CRC), gastric cancer (GC), and other cancers [5, 6]. In CRC and GC, frame-shift mutations in TGFBR2 leading to MSI formation are more prevalent, suggesting that specific tumor environments may contribute to MSI occurrence [4, 5].
Numerous studies have examined mutation profiles in solid tumors, but the somatic mutations of cancer-associated genes have not been explored based on the MSI status. This study aimed to investigate the MSI status of patients with solid organ tumors and identify somatic gene variations in these patients.
Material and methods
Ethics and study subjects
This study, approved by the Institutional Non-Interventional Ethics Committee (2023/87) following the Helsinki Declaration, retrospectively examined the records of 192 patients diagnosed with solid organ cancer aged 18 and over. These patients, referred to the Molecular Pathology Laboratory from the Oncology Clinic of Aydın Adnan Menderes University Medical Faculty Hospital between January 2018 and December 2022, were screened for eligibility. Excluded were cases with undetermined diagnoses. Data, including age, gender, and diagnostic subgroups at the time of diagnosis, were extracted from the Molecular Pathology Laboratory’s electronic database. The collected data were then categorized based on age and clinicopathological characteristics for the NGS colon cancer mutation panel, real-time polymerase chain reaction (PCR) analysis for MSI, and immunohistochemical (IHC) MMR protein analysis.
DNA isolation
DNA isolation was performed from 10 μm thick sections of the patients’ paraffin-embedded tissues using the FFPE DNA isolation kit (GeneReader FFPE kit, Qiagen, Hilden, Germany) following the kit procedure. The purity and quantity of the isolated DNA samples were determined using the Qubit 3.0 fluorometer and Qubit 3.0 dsDNA HS assay kit (Life Technologies, California, USA). DNA samples yielding at least 100–150 ng were used to continue the study.
MSI detection using real-time PCR
MSI detection was performed using the EasyPGX Ready MSI kit based on the denaturation profile with real-time PCR. The microsatellite markers BAT25, BAT26, NR21, NR22, NR24, NR27, CAT25, and MONO27, which are recommended by the National Cancer Institute for MSI detection, were investigated. MSI detection was performed by comparing the MS loci identified in the tumor tissues of patients with solid organ tumors to the microsatellite loci in the normal tissues of the same patient. The instability of one locus was identified as MSI-L; MSI-H, on the other hand, refers to the instability of two or more loci, while MSS indicates stability in all five loci.
Detection of MMR protein expression by immunohistochemistry (IHC)
Since the detection of MMR gene deletions indirectly reflects MSI, the expression of MMR protein arising from hMLH1, hMSH2, hMSH6, and PMS2 was detected using IHC. In brief, 3 μm thick FFPE tissue sections were paraffinized in xylene, rehydrated in a series of graded alcohols, washed with double-distilled water, and subjected to pre-treatment with DAKO solution (EnVision FLEX Target Retrieval Solution, High pH 50x) at 97°C. Later, the slides were subjected to incubation with primary monoclonal antibodies targeting MLH1, PMS2, MSH2 and MSH6 for 30 min. The analyses were performed on the automatic platform Autostainer Link 48 (Dako, Carpinteria, CA, USA) following the manufacturer’s protocol. The interaction between the antigen and antibody was observed using diaminobenzidine as the chromogen with the EnVision FLEX kit. In the four IHC stainings performed, nuclear staining in both the internal control tissue and tumor tissue was evaluated to determine whether there was nuclear staining loss. The absence of expression of any of these MMR proteins is referred to as MMR-deficient (dMMR), while the presence of all four MMR proteins is considered MMR-proficient (pMMR). In our analysis, dMMR is considered equivalent to MSI-H.
NGS multiple gene variation analysis
It is believed that specific tumor environments with gene variations are conducive to the formation of MSI. Therefore, in MSI assessments, it is important to evaluate multiple gene variations, including MMR genes. As a result, the colon cancer NGS panel (Colon cancer panel DHS-002Z, Qiagen, Hilden, Germany) (Table I) test was applied to the patients. After isolating the DNA, the NGS workflow progressed through several stages, encompassing target enrichment, library preparation, template preparation, sequencing, variant calling, variant classification, and the interpretation of the resulting data. The sequencing procedure was conducted utilizing a MiniSEQ NGS platform (MiniSEQ, MN00676) with Illumina, Inc.’s MiniSEQ High Output Reagent Cartridge (San Diego, CA, USA).
Table I
Gene list in colon cancer NGS panel
NGS variant assessment and statistical analysis
The NGS variant analyses were conducted by combining pathological and clinical findings with the automatic bioinformatics support provided by Qiagen Clinical Insight Interpret 8.1.202021. Based on their significance to cancer diagnosis, prognosis, and/or therapeutic implications, the variants were categorized into four tiers (Tiers I-IV). Variants with strong clinical significance were categorized as Tier I and Tier II. Those with unknown clinical significance due to insufficient evidence were labeled as Tier III, while variants with enough evidence to be classified as benign or possibly benign were categorized as Tier IV.
Results
The patient cohort included 108 cases of CRC, 19 GC, 13 pancreatic cancer (PC), 2 small intestine cancer (SIC), 6 endometrial cancer (EC), 8 ovarian cancer (OC), 3 bladder urothelial cancer (BUC), 2 cervical cancer (CC), and 31 cases of lymph node and distant organ metastasis (liver, lung, brain). Among CRC cases, 10 had lymph node metastasis, 12 had liver metastasis, 8 had lung metastasis, and 1 case had brain metastasis. Regarding GC cases, only 1 case had liver metastasis. In the total cohort of 192 patients, 99 (51.6%) were male, and 93 (48.4%) were female, with ages ranging from 25 to 87 years and an average age of 56 years.
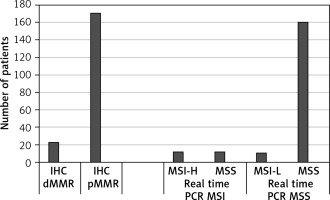
In the IHC assessment, 22 (11.45%) cases were identified as dMMR (MSI-H), while 170 (88.55%) cases were pMMR. Among the 22 dMMR cases identified by real-time PCR, 11 exhibited MSI-H, and 11 showed MSS. Out of the 170 pMMR cases, 160 were MSS according to real-time PCR, and 10 cases exhibited MSI-L (Figure 1). Specifically, instability was observed in 2 cases for MONO27, 6 cases for BAT25, 1 case for BAT26, and 1 case for NR22 in the MSI-L cases. In summary, only 22 (11.45%) cases demonstrated discordant results between real-time PCR and IHC MSI analyses.
In CRC patients, MSI-H was present in 5 (4.62%) cases, in EC patients in 3 (50%) cases, and in GC patients in 1 (5.26%) case. Among CRC cases with MSI-H, 2 (40%) cases showed lymph node metastasis, with no distant organ metastases. In other cancer cases with MSI-H, neither lymph node nor distant organ metastasis was observed.
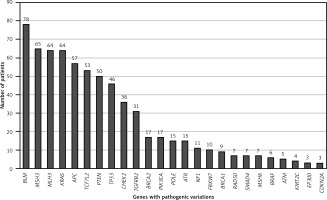
The average age of patients with MSI-positive tumors was 56.63, compared to 53.08 for patients with MSS tumors. However, this difference was not statistically significant (p = 0.067). Among MSS cases, 34 (51.5%) were male, and 32 (48.5%) were female. For MSI cases, 13 (56.5%) were male, and 10 (43.5%) were female. The gender distribution difference between MSI and MSS cases did not reach statistical significance (p = 0.09). All cases exhibited multiple variations, and the variation rates identified in our study are illustrated in Figure 2.
According to NGS analysis, the top four pathogenic variants observed in all cases are as follows: BLM exon 7 c.1544delA (40.62%, 78/192), MSH3 exon 7 c.1148delA (33.85%, 65/192), MLH3 exon 2 c.1755delA (33.33%, 64/192), and KRAS exons 2-4 (33.33%, 64/192). Additionally, the following pathogenic variants were observed at lower percentages: APC exons 2, 6, 9, 10, 14-16 (29.68%, 57/192), TCF7L2 exon 14 (27.60%, 57/192), PTEN exons 1, 4-8 (26.04%, 50/192), TP53 exons 5-8, 10 (23.96%, 46/192), CHEK2 exons 4, 13-14 (18.75%, 36/192), and TGFBR2 exon 3 (16.15%, 31/192). BLM mutations were most commonly observed in conjunction with variations in other genes.
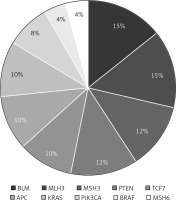
In all cancer cases with MSI-H, the most frequently observed pathogenic somatic variations include BLM, cancer driver genes (APC, KRAS, TP53), DNA repair genes (MLH3, MSH3), PTEN, TCF7L2, and PIK3CA variations. The pathogenic variations identified in MSI-H cases are summarized in Figure 3. MSI-H cancer patients exhibited a higher burden of variations compared to MSS cancer patients.
In all CRC cases with MSI, the three most frequently observed pathogenic variants were BLM exon 7 c.1544delA (46.30%, 50/108), MSH3 exon 7 c.1148delA (41.66%, 45/108), and MLH3 exon 12 c.1755delA (41.66%, 45/108). A similar pattern was observed in overall CRC somatic variant profiles in MSI cases, with BLM exon 7 c.1544delA (100%, 5/5), MSH3 exon 7 c.1148delA (100%, 5/5), MLH3 exon 12 c.1755delA (80%, 4/5), APC exons 10, 14, 16 variations (80%, 4/5), and TCF7L2 exon 14 c.1403delA (80%, 4/5) being the most frequent pathogenic variants. In MSS cases, a statistically significantly higher frequency of certain variants was observed compared to MSI cases, including BLM exon 7 c.1544delA, MSH3 exon 7 c.1148delA, MLH3 exon 12 c.1755delA, KRAS exon 2 c.35G-A, APC exon 16 variations, TCF7L2 exon 14 c.1403delA, PTEN exon 5 c.407G-A, TGFBR2 exon 3 c.383dupA, CHEK2 exon 14 c.1556C-T, and POLE exon 34 c.4337_4338delTG (p = 0.02). In MSI-positive patients, the following variant combinations of IHC marker proteins were observed; (1) 1 patient had a combination of PMS2 exon 11 c.1239delA and MSH6 exon 5 c.3261dupC variants, (2) 1 patient had a combination of MLH1 exon 8 c.676C-T and MSH6 exon 4 c.2314C-T variants, and (3) 2 patients had an MSH6 exon 5 c.3261dupC variant. In other MSI-positive cases, no variants related to IHC marker proteins were detected. In MSS cases, 1 patient had the MSH6 exon 5 c.3261dupC variant.
The most commonly observed pathogenic variants in GC patients were BLM exon 7 c.1544delA (21.05%, 4/19), MSH3 exon 7 c.1148delA (21.05%, 4/19), MLH3 exon 12 c.1755delA (15.78%, 3/19), TCF7L2 exon 14 (15.78%, 3/19), and TGFBR2 exon 3 c.383dupA (15.78%, 3/19). In MSI-positive GC patients, BLM exon 7 c.1544delA, MSH3 exon 7 c.1148delA, and TCF7L2 exon 14 c.1403delA variants were observed, while in MSS cases, the somatic variation profile observed in all GC patients was seen.
The most common pathological variants in EC cases were PTEN exon 5 c.407G-A, exon 6 c.600delT, and exon 7 c.645dupT (83.33%, 5/6), BLM exon 7 c.1544delA (83.33%, 5/6), PIK3CA exon 2 c.263G-A, exon 10 c.306A-T, and exon 21 c.3140A-G (66.66%, 4/6), MLH3 exon 12 c.1755delA (50%, 3/6), CHEK2 exon 14 (50%, 3/6), and MSH3 exon 7 c.1148delA (33.33%, 2/6). In MSI-positive EC cases, variants included PTEN exon 6 c.600delT, PIK3CA exons 2, 10, and 21, BLM exon 7 c.1544delA, and CHEK2 exon 14. In all MSS EC cases, common variants were BLM exon 7 c.1544delA, MLH3 exon 12 c.1755delA, PTEN exon 5 c.400G-A, CHEK2 exon 14 c.1556C-T, TCF7L2 exon 14 c.1385delA, MSH3 exon 7 c.1148delA, and BRCA1 exon 11 c.1961delA. MSI-detected EC patients had 1 case with the variant PMS2 exon 11 c.1239delA related to IHC markers.
In PC cases, frequent variants included TCF7L2 exon 14 c.1403delA (30.77%, 4/13), MLH3 exon 12 c.1755delA (30.77%, 4/13), MSH3 exon 7 c.1148delA (23.07%, 3/13), and PTEN exon 5 c.407G-A (15.38%, 2/13). In OC cases, ATR exon 10 c.2320del (62.5%, 5/8), CHEK2 exon 14 c.1556C-T (50%, 4/5), and BLM exon 7 c.1544delA (37.50%, 3/5). BUC patients had BLM exon 7 c.1544delA (33.33%, 1/3), MSH3 exon 7 c.1148delA (33.33%, 1/3), TCF7L2 exon 14 c.1403delA (33.33%, 1/3), and TCERG1 exon 18 c.2807delA (33.33%, 1/3). CC patients exhibited BLM exon 7 c.1544delA (33.33%, 1/3), MSH3 exon 7 c.1148delA (33.33%, 1/3), TCF7L2 exon 14 c.1403delA (33.33%, 1/3), MLH3 exon 12 c.1755delA (33.33%, 1/3), and TP53 exon 8 c.796G-A (33.33%, 1/3). SIC patients had BLM exon 7 c.1544delA (50%, 1/2), MSH3 exon 7 c.1148delA (50%, 1/2), TGFBR2 exon 3 c.383dupA (50%, 1/2), KRAS exon 2 c.35G-T (50%, 1/2), TP53 exon 5 c.400T-C (50%, 1/2), and APC exon 16 c.1544delA (50%, 1/2) variants.
In CRC patients with lymph node metastasis, common variants were BLM exon 7 c.1544delA (20%, 2/10), MSH3 exon 7 c.1148delA (20%, 2/10), MLH3 exon 12 c.1755delA (20%, 2/10), KRAS exon 2 c.35G-T (20%, 2/10), and exon 4 c.436G-A (20%, 2/10). In CRC patients with lymph node metastasis and MSI, 1 patient had PMS2 exon 11 c.1239delA and MSH6 exon 5 c.3261dupC variants in IHC marker proteins. Lung metastasis cases showed frequent somatic variations in PIK3CA exon 8 c.1357G-C and exon 21 c.3073A-G, as well as TP53 exon 5 c.407G-A and exon 10 c.1015G-T variants. Brain metastasis cases exhibited CHEK2 exon 14 c.1556C-T variation. Liver metastasis cases had variations in KRAS exon 2 c.38G-A, exon 3 c.182A-T, and exon 4 c.436G-A, as well as TP53 exon 5 c.395A-G, exon 7 c.712T-C, and exon 8 c.785G-T, and PTEN exon 5 c.407G-A.
Discussion
In CRC treatment, immune checkpoint inhibitors have proven effective in tumors with MSI-H/dMMR status [5]. Hence, it is vital to assess MSI-H/dMMR status in diverse solid organ tumors. In our study, it was deemed essential to evaluate the MSI-H/dMMR status in different solid organ tumors and analyze the somatic variation profiles based on MSI status [6].
Two main diagnostic methods are used for detecting MSI in cancer, involving molecular techniques for MSI assessment and IHC to gauge the expression of four MMR proteins on histological tissue sections. However, documented discordance exists between these methods [7–9]. IHC, widely available in pathology laboratories, identifies deficient MMR proteins by comparing expression levels in tumor cells and adjacent normal tissue [10, 11]. Approximately 6–7% of MSI tumors show ambiguous IHC results, particularly with complex factors such as mutated MMR proteins, POLE mutations, or MLH1 promoter methylation [12, 13]. In such cases, ESMO guidelines recommend molecular testing for MSI detection [14]. The MSI test, based on PCR amplification of microsatellite markers, aligns strongly (about 90–95%) with MMR-IHC. Despite IHC being the established method for MSI biomarker analysis, our study suggests that real-time PCR platforms can effectively determine MSI status, producing results akin to IHC [8–11, 15]. A high concordance rate (approximately 88.55% for MSS/pMMR and MSI-H/dMMR cases) was evident between the real-time PCR platform and IHC, consistent with existing literature.
Among the 22 discordant cases, five different tumor types and two distant organ metastases exhibited distinct IHC and real-time PCR profiles. These included 13 CRCs, two PCs, one CC, one EC, one BIC, three liver metastases, and one lung metastasis. Diverse molecular mechanisms underlie MSI in various organs, potentially impacting MSI testing and MMR IHC outcomes. For instance: (1) MSH6-negative tumors might manifest as MSS or MSI-L in MSI analysis. (2) Tumors with direct mutations can display an MSI profile due to microsatellite locus mutation accumulation, irrespective of preserved MMR protein expression. (3) About 3% of EC cases exhibit subclonal MMR protein expression loss, with subtle microsatellite shifts characterizing MSI in EC, posing challenges for diagnostic interpretation. (4) The utilization of outdated IHC evaluation criteria can influence MMR assessment. (5) Distinct PCR-based approaches are influenced by inherent limitations that adequate user training and high-quality samples can address [16].
In a prior report, high dMMR rates were noted in around 15% of early-stage CRC, 5% of advanced-stage CRC, 20–30% of EC, and 28.9% of GC, followed by PC and OC at 7.5% and 5.3%, respectively [17]. Our study observed 4.62% of CRC patients, contrasting with the literature, with 50% of EC patients and 5.26% of GC patients having dMMR/MSI-H status, possibly influenced by differing ethnic profiles.
MSI and MMR status agreement in metastatic cancer may vary by organ. Consistency is usually strong in liver, lung, and distant lymph node metastases, but discrepancies are more likely in the peritoneum or ovaries [18]. In our study, two CRC cases (40%) with MSI-H were concordant with lymph node metastasis, while distant organ metastases were absent. No lymph node or distant organ metastases were observed in other MSI-H cancer cases.
In a specific organ, MSI-H cancers typically share more genomic mutations with MSS counterparts than with MSI-H tumors in other organs. These cancers show shared mutations in MS loci and are marked by elevated mutation loads in coding and non-coding regions, featuring frame-shift mutations from indels and a lower count of single-nucleotide mutations [19]. Le et al. found that pMMR cancer patients (n = 6) had an average of 73 mutations per tumor (p = 0.007), while dMMR cancer patients (n = 9) had an average of 1782 somatic mutations per tumor [20]. Consistent with the literature, MSI-H cancer cases in this study also showed a higher variation burden, indicating that MSIs contribute to the genetic diversity and complexity of solid tumors.
The absence of MMR genes in tumor cells, lack of protein synthesis, or errors during the replication repair process leads to DNA-MMR deficiency and MSI. As a result, genes that are essential for DNA replication and repair or apoptosis control are frequently mutated in MSI cancers. These genes include MMR genes, MED1, ATM, DNA helicase, BLM, E2F4, RIZ, and caspase-5 genes [21]. In our study, the most commonly observed variations in the presence of MSI in all solid organ cancers were in the DNA replication-related BLM gene and in genes involved in DNA repair with pathogenic variations.
In Chinese cohort studies focused on CRC, it was observed that cancer driver genes such as APC, TP53, and KRAS exhibited frequent mutations in CRC samples, irrespective of the MSI status. However, MSI-H cases exhibited a significantly higher number of alterations compared to MSS cases [22, 23]. Among MSI-H cases, the most prevalent mutation identified was TCF7L2. This type of frame-shift mutation in the TCF7L2 gene has been notably observed in high MSI CRC and GC. TCF7L2 holds significance as a key member of the WNT signaling pathway. Additionally, it is worth noting that mutations in genes associated with the WNT pathway have shown enrichment in MSI-H cases, as evidenced by a study involving a broad cohort of 67 000 pan-tumor samples. The MMR genes in MSI-H samples were found to undergo a high rate of mutations, indicating that MSI is a consequence of dysfunction in the MMR gene function [24]. In our study, it was found that TCF7L2 gene variations in MSI-H CRC patients occurred at a rate of 80%, which is consistent with the literature, along with APC and DNA repair gene variations.
According to previous reports in the literature conducted on the Korean population, TP53 mutations were the most common (35%) in MSI-H GC cases, followed by EGFR (8%), HNF1A (8%), PIK3CA (8%), and ERBB2 (5%) mutations [24]. In our study, we observed more frequent variations in the BLM, MSH3, and TCF7L2 genes due to population differences.
Statistically significant mutation rates are evident in MSI-H EC, with notable frequencies of mutations observed in various genes. The most notable mutation frequencies include PTEN (88%), PIK3CA (54%), PIK3R1 (41%), RPL22 (37%), ARID1A (37%), KRAS (35%), and ZFHX3 (31%) [25]. In our study, we also observed the highest percentage of variations in PTEN and PIK3CA among MSI-H EC cases.
Overall, our data indicate that MSI-H solid organ tumors harbor more somatic mutations compared to those that are MSS/MSI-L. These findings suggest that the accumulation of MSI contributes to the genetic diversity and complexity of solid organ tumors. Indeed, since solid organ tumors exhibit distinct patterns based on ethnic origin, further studies involving different racial/ethnic groups or cancer types can strengthen our research findings.
This study had several limitations. To begin with, it was a retrospective study carried out within a singular institution and involving a single ethnic group. As a result, the potential to apply the findings to other populations could be restricted. Secondly, the analysis of MSI was performed by IHC and real-time PCR, but more advanced molecular methods such as NGS could potentially provide more accurate results.