Introduction
Gallbladder cancer (GBC) is a relatively rare type of cancer, yet it accounts for more than 80 percent of all biliary tract malignancies [1, 2]. Despite great improvement in clinical GBC treatment, most GBC patients are usually diagnosed at advanced stages with extremely ominous outcomes [3]. To date, our understanding regarding molecular mechanisms of GBC remains unclear. Therefore, this study aimed to investigate the mechanisms underlying GBC development and metastasis, in order to develop novel therapeutic strategies and improve prognosis for GBC patients.
MicroRNAs (miRNAs/miRs), a class of small (~22 nucleotides) non-coding RNAs, bind directly to the 3′-untranslated regions (3′UTRs) of their target mRNAs to stimulate mRNA degradation or inhibit protein translation [4, 5]. Accumulating research has reported that miRNAs serve as tumor modulators that might be utilized as powerful diagnostic markers or therapy targets for cancer diagnosis and treatment [4, 6–9]. Moreover, recent studies have demonstrated that a large number of miRNAs exhibit regulatory roles in the initiation and progression of GBC, such as miR-31, miR-33a, miR-1, miR-145 and miR-26a [6, 10–12].
miR-520b is proved to be a tumor suppressor in various types of human tumors, including colorectal cancer, osteosarcoma, glioma and head-neck cancer [7, 13–15]. In contrast, Guan et al. reported that miR-520b stimulated ovarian cancer proliferation [16]. However, the effects of miR-520b on GBC remain unclear. In our study, we discovered that miR-520b was down-regulated in human GBC tissues. Hence, we hypothesized that miR-520b might have suppressive effects on GBC development. To verify this hypothesis, NOZ cells were employed to investigate the role of miR-520b in GBC.
Material and methods
Patients and tissue specimens
This study was approved by the ethics committee of The First Hospital of Jilin University. Informed consent was provided from all 30 patients. Tumor tissues and neighboring normal tissues were harvested from GBC patients after radical tumor resection.
MiRNAs, plasmid construction and transfection
MiR-520b mimic (miR-520b mimic), negative control of miR-520b mimic (NC mimics), miR-520b inhibitor (miR-520b inhibitor) and negative control of miR-520b inhibitor (NC inhibitor) were bought from GenePharma (Shanghai, China). RAB22A cDNA was inserted into a pCMV expression vector (LQbiotech, Shanghai, ChinaEra Biotech) to construct the RAB22A overexpression vector. MiRNA or plasmid transfection was performed using Lipofectamine 2000 (Invitrogen, Beijing, China).
Cell culture
The NOZ cell line was purchased from Shanghai Cell Institute Country Cell Bank (Shanghai, China) and maintained in high-glucose Dulbecco’s Modified Eagle’s Medium (DMEM, Life Technologies, Beijing, China) supplemented with 10% fetal bovine serum (Life Technologies, Beijing, China) at 37°C.
MTT assay
Ten thousand transfected NOZ cells were seeded and incubated in 12-well plates for 48 h. Then, 0.5 mg/ml MTT was added to each well. After 4 h, formazan crystals were dissolved in DMSO and OD 490 nm was measured.
Colony formation assay
Two hundred transfected NOZ cells were plated and incubated in 6-well plates for 48 h. After fixation, the colonies were dyed using crystal violet. Images were taken and colonies were counted.
Wound healing assay
One hundred thousand transfected NOZ cells were plated in 24-well plates. When GBC cells reached around 80% confluency, the cell layer was scraped using a 200 μl pipette tip. Images of the wounded areas were taken 24 h after treatment via an inverted light microscope.
Transwell invasion assay
One hundred thousand transfected NOZ cells were suspended in 500 μl of DMEM medium without FBS and transferred into Matrigel-coated upper chambers. 500 μl of complete medium was supplemented into the lower chambers. 48 h after seeding, residual cells were removed and invasive cells were fixed, dyed and counted under a microscope.
Quantitative real-time PCR
Total RNA was extracted from frozen tissue samples or NOZ cells, and then transcribed into cDNA via a reverse transcription kit (TaKaRa, Beijing, China). RNA level was examined through quantitative real-time PCR (qPCR) using SYBR Green (Takara, Beijing, China). Primers used in this study were as follows:
miR-520b-fwd: 5’-AACACGCAAAGTGCTTCCTTTTAGA-3’
miR-520b-rev: 5’-CAGTGCAGGGTCCGAGGT-3’
RAB22A-fwd: 5’-GTGTGTCTGCTCGGGGATAC-3’
RAB22A-rev: 5’-GCCCCTATTGTTGGGTTGATGT-3’
β-Actin-fwd: 5’-GGAGATTACTGCCCTGGCTCCTA-3’
β-Actin-rev: 5’-GACTCATCGTACTCCTGCTTGCTG-3’
Immunohistochemical (IHC) staining
GBC tumor tissues and neighboring normal tissues were fixed, embedded, dewaxed and hydrated. After blocking, the sections were immersed with the primary antibody against RAB22A (Abcam, Beijing, China). After washing, sections were probed with the secondary antibodies for 1 h at 22°C. Sub-sequently, the sections were stained via DAB for 8 min and counterstained via hematoxylin for 2 min. Images were taken using a fluorescence microscope.
Western blot analysis
Protein lysates of frozen tissue or cells were prepared. After quantification, proteins were separated by SDS-PAGE and then transferred onto PVDF membranes. After blocking, PVDF membranes were probed via primary antibody against RAB22A (1 : 500; Abcam, Beijing, China); anti-GAPDH antibody (1 : 3000; Abcam, Beijing, China) was taken as the internal reference. Then, after incubation with secondary antibodies, the protein bands were quantified by an ECL chemiluminescence kit.
Dual luciferase reporter assay
Wide-type RAB22A 3′UTR (containing putative miR-520b binding sites) or mutant RAB22A 3′UTR (harboring the mutant binding sequences of miR-520b) was generated and inserted into a luciferase reporter plasmid (LQbiotech, Shanghai, China) to generate wild-type (RAB22A 3′UTR-WT) or mutant report plasmid (RAB22A 3′UTR-MUT) respectively.
miR-520b mimic, as well as RAB22A 3′UTR-WT or RAB22A 3′UTR-MUT, was transfected into NOZ cells. 48 h after transfection, luciferase reporter assay was conducted using a luciferase assay kit (Promega, Shanghai, China).
Statistical analysis
Data were analyzed using SPSS 18.0 software and presented as mean ± SD. Comparisons between two groups were assessed via one-way ANOVA. The correlation between miR-520b and RAB22A expression levels was examined via Pearson’s χ2 test. A difference was considered statistically significant when p < 0.05 in this study.
Results
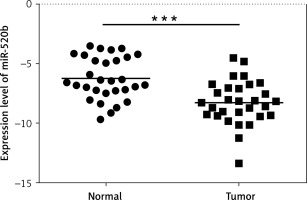
MiR-520b was down-regulated in GBC tumor tissues. qPCR assay was employed to investigate the effects of miR-520b on GBC development. Figure 1 reveals that miR-520b RNA level in tumor samples was significantly lower than that in the neighboring normal tissues (p < 0.001).
Figure 1
miR-520b was down-regulated in GBC tumor tissue samples. qPCR analysis of miR-520b mRNA expression in the tumor tissues and neighboring normal tissues from GBC patients (mean ± SD)
*p < 0.05, **p < 0.01, ***p < 0.001.
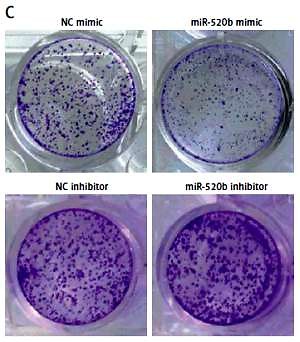
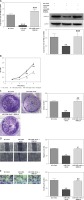
MiR-520b inhibited the proliferation of NOZ cells
qPCR confirmed there was an increase of miR-520b mRNA in miR-520b mimic groups compared to that in the NC mimic group (Figure 2 A, p < 0.001). In contrast, miR-520b inhibitor significantly decreased miR-520b mRNA level in NOZ cells in comparison to the NC inhibitor group (Figure 2 A, p < 0.01). MTT assay showed that miR-520b mimic inhibited the proliferation in NOZ cells time-dependently (Figure 2 B, p < 0.01). However, Figure 2 B demonstrates that down-regulation of miR-520b facilitated the proliferation of NOZ cells in a time-dependent manner (p < 0.01). In addition, colony formation assay indicated miR-520b mimic overexpression NOZ cells had a much lower number of colonies in comparison with those in the NC mimic group (Figure 2 C, p < 0.01), whereas miR-520b inhibitor stimulated NOZ colony formation compared to NC inhibitor (Figure 2 C, p < 0.05).
Figure 2
miR-520b inhibited NOZ cell proliferation. NOZ cells were transfected with miR-520b mimic or miR-520b inhibitor. A – MiR-520b mRNA level in transfected NOZ cells was determined by qPCR. B, C – Cell viability of transfected NOZ cells was determined via MTT assay (B) and colony formation assay (C) (mean ± SD)
*p < 0.05, **p < 0.01, ***p < 0.001.
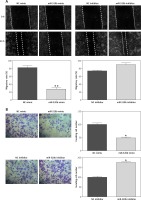
MiR-520b restricted the migration and invasion of NOZ cells
Figure 3 A shows that miR-520b mimic inhibited the migration of NOZ cells (68.09%, p < 0.01) compared to NC mimic, whereas miR-520b inhibitor exhibited no significant influence on the migratory capability of NOZ cells. Additionally, miR-520b mimic limited the invasion ability of NOZ cells (Figure 3 B, 52.94%, p < 0.05), whereas miR-520b inhibitor fostered their invasion capacity (Figure 3 B, 90.91%, p < 0.05).
RAB22A was a direct downstream target of miR-520b
Bioinformatics analysis using TargetScan revealed that there was a potential miR-520b binding site in the RAB22A 3′UTR (Figure 4 A). Dual luciferase reporter assay further revealed that miR-520b mimic decreased the luciferase activity of wild-type RAB22A 3′UTR in comparison to NC mimic (Figure 4 A, 55.56%, p < 0.01). However, miR-520b mimic exhibited little effect on the luciferase activity of mutant RAB22A 3′UTR (Figure 4 A). In addition, Figure 4 B shows that NOZ cells transfected with miR-520b mimic had a significantly lower RAB22A protein level in comparison to cells transfected with NC mimic (p < 0.001). However, miR-520b inhibitor stimulated RAB22A expression compared to the NC inhibitor group (Figure 4 B, p < 0.05).
Figure 4
RAB22A was a direct target of miR-520b. A – Schematic diagram of miR-520b binding sites in RAB22A 3′UTR. Luciferase reporter activities driven by wild-type or mutant RAB22A 3′UTR were examined in NOZ cells transfected with miR-520b mimic or NC mimic. B – RAB22A protein and mRNA level in NOC cells transfected with miR-520b mimic or miR-520b inhibitor were examined via Western blot and qPCR, respectively. β-actin was used as an internal control for qPCR assay (mean ± SD)
*p < 0.05, **p < 0.01, ***p < 0.001.
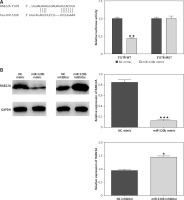
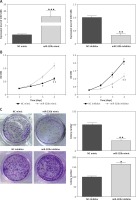
RAB22A overexpression blocked the anti-tumor effects of miR-520b on NOZ cells
RAB22A overexpression restored the inhibition of RAB22A mRNA and protein expression in NOZ cells caused by miR-520b mimic (Figure 5 A, p < 0.001). MTT assay showed that miR-520b mimic time-dependently decreased the viability of NOZ cells in comparison with NC mimic (Figure 5 B, p < 0.01), whereas RAB22A overexpression completely abolished this miR-520b mimic-induced reduction of viability (Figure 5 B, p < 0.01). Colony formation assay proved that miR-520b mimics significantly inhibited NOZ cell proliferation in comparison with NC mimic, whereas RAB22A overexpression inhibited the above phenomenon (Figure 5 C, p < 0.01). Additionally, wound healing assay demonstrated that miR-520b mimic inhibited GBC cell migration by ~50% compared to NC mimic, whereas the inhibitory effect of miR-520b on GBC migration could be restored upon RAB22A overexpression (Figure 5 D, p < 0.05). Transwell invasion assay also showed that RAB22A overexpression restricted the inhibitive effect of miR-520b on the invasion ability of NOZ cells (Figure 5 E, p < 0.01).
Figure 5
RAB22A overexpression blocked the anti-tumor effects of miR-520b on NOZ cells. NOZ cells were transfected with NC mimic, miR-520b mimic, or miR-520b mimic with RAB22A overexpression plasmid (miR-520b mimic + RAB22A). A – RAB22A mRNA and protein levels were detected via qPCR and Western blot, respectively. B – The viability of transfected NOZ cells was detected via MTT assay. C – The viability of transfected NOZ cells was detected via colony formation assay. D, E – The migration and invasion of transfected NOZ cells were investigated via wound healing (D) and transwell invasion assay (E) respectively (mean ± SD)
*p < 0.05, **p < 0.01, ***p < 0.001 compared to NC mimic group; #p < 0.05, ##p < 0.01, ###p < 0.001 compared to miR-520b mimic group.
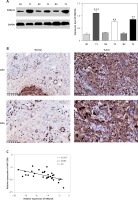
RAB22A was up-regulated in GBC tumor tissues and had a negative relationship with miR-520b expression
Western blot analysis indicated that tumor samples of GBC patients had a higher RAB22A protein expression level (p < 0.001, p < 0.01 and p < 0.01 for T1, T2 and T3 compared to matched neighboring normal tissues respectively) than that in matched neighboring normal tissues (Figure 6 A). Also, IHC analysis showed that RAB22A expression was enhanced in GBC tumor samples compared to neighboring normal tissues (Figure 6 B). Moreover, Figure 6 C further confirmed that there was a negative regulatory relationship between miR-520b and RAB22A.
Figure 6
RAB22A was up-regulated and had negative relationship with miR-520b expression in GBC tumor tissues. A – Western blot analysis of RAB22A expression in tumor tissues (T) and neighboring normal tissues (N) from 3 GBC patients. GAPDH was used as the internal loading control. B – IHC staining of RAB22A in GBC tumor sample and neighboring normal tissue sample. C – Correlation of RAB22A and miR-520b expression in GBC tumor samples (mean ± SD)
**p < 0.01, ***p < 0.001.
Discussion
Gallbladder carcinoma, the most common and lethal biliary neoplasm, is associated with a poor survival rate due to late clinical diagnosis and its highly malignant properties, including rapid development and frequent occurrence of early metastasis. Nowadays, many studies have proved that miRNA aberrations are associated with GBC progression. For instance, Zhou et al. reported that miR-26 expression was decreased in human GBC tissues in comparison with that in matched neighboring normal tissues, and miR-26 might show tumor suppressive effects on GBC [6]. According to Zhang et al., miR-33a was down-regulated in human GBC tissues, which functioned as a tumor suppressor in GBC by targeting Twist [12]. Jin et al. found that miR-143-3p was down-regulated in GBC tumor tissues and was an inhibitor of GBC progression [17]. Hence, miRNAs may be used as therapeutic markers for GBC [18].
In this study, we found that miR-520b was significantly down-regulated in GBC patients’ tissues compared to neighboring normal tissues. Moreover, miR-520b mimic significantly inhibited the viability, colony formation ability, migration and invasion rates of NOZ cells. Conversely, miR-520b inhibitor decreased miR-520b expression and subsequently fostered the above malignant phenotypes of NOZ cells. Therefore, miR-520b had tumor suppressive activities against GBC development in vitro.
miRNAs could modulate mRNA expression of target genes by binding to the 3′UTR of target genes. Previous studies have revealed several targets of miR-520b in various tumors, such as MBD2 in glioma [13], CD44 in head-neck cancer [7], DCUN1D1 in colorectal cancer [15] and TET1 in hepatocellular carcinoma [19]. In this study, bioinformatics analysis predicted that RAB22A 3′UTR was a potential binding site for miR-520b.
RAB22A, a member of the RAB family, is a small GTPase. Many studies have proved that RAB22A is often upregulated and acts as an oncogene in various malignant tumors. For example, Zhou et al. reported that RAB22A played an oncogenic role in lung cancer migration and invasion in vitro [20]. Su et al. reported that high expression of RAB22A facilitated melanoma growth [21]. Xiong et al. reported that RAB22A overexpression fostered the progression of renal cell carcinoma [22]. In addition, increasing evidence has suggested that RAB22A was a direct target of several miRNAs in different types of tumors, such as miR-211 in melanoma [23], miR-373 in ovarian cancer [24], miR-203 in osteosarcoma [25] and miR-204 in renal cell carcinoma [22]. Dual luciferase reporter assay indicated that miR-520b mimic significantly restricted the luciferase activity of the wild type RAB22A 3′UTR, whereas the luciferase activity of mutant RAB22A 3′UTR was not changed upon miR-520b mimic overexpression. Collectively, the above results confirmed that miR-520b could bind to the RAB22A 3′UTR in NOZ cells. Furthermore, Western blot and qPCR assay confirmed that RAB22A mRNA and protein expression were negatively associated with miR-520b overexpression in NOZ cells.
Based on our findings, we hypothesized that the anti-tumor roles of miR-520b in NOZ cells may be induced by the modulation of RAB22A overexpression. To prove our hypothesis, we transfected NOZ cells with or without RAB22A upon via miR-520b mimic overexpression. A series of cellular experiments indicated that RAB22A overexpression eradicated the suppressive effects of miR-520b on the viability, colony formation, migration and invasion abilities of NOZ cells. In addition, we observed higher levels of RAB22A mRNA and protein in human GBC tissues than in corresponding normal tissues, suggesting that RAB22A expression was positively associated with GBC progress. Moreover, RAB22A expression was proved to be negatively linked to miR-520b expression in human GBC tissues, which was consistent with our results of NOZ cells.
In conclusion, this study confirmed that miR-520b exhibited anti-tumor effects on the proliferation, migration and invasion abilities of GBC cells by targeting RAB22A for the first time, which not only presented novel mechanisms underlying GBC development but also provided therapeutic targets for GBC patients. Increasing evidence shows that miRNA replacement therapy (the improvement of miRNA activity) may regain the lost function and is a very promising cancer treatment method [26, 27]. Hence, our findings offered a rationale to study “miR-520b replacement therapy during GBC treatment”. However, in vivo delivery of miRNA is still a big challenge for the clinical application of miR-520b replacement therapy. Moreover, further experiments still need to be performed to analyze the anti-tumor effects of miR-520b on GBC in vivo, with the aim of developing novel therapeutic strategies from bench to clinic.
In conclusion, MiR-520b showed protective effects on GBC by targeting RAB22A, as shown by significantly blocking the proliferation, migration and invasion abilities of NOZ cells.