Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Section Editors
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
IMMUNOLOGY / CLINICAL RESEARCH
Kidney microbiome in patients with kidney carcinoma: role of SA and SNZ gene expression
1
Department of Urology, The First Affiliated Hospital of Anhui Medical University, Anhui, China
2
Institute of Urology, Anhui Medical University, Hefei, Anhui, China
3
Anhui Province Key Laboratory of Genitourinary Diseases, Anhui Medical University, Hefei, Anhui, China
4
Department of Oncology, The Fourth Affiliated Hospital of Anhui Medical University, Hefei, Anhui, China
5
Institute of Transfusion Medicine and Immunology, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany
Submission date: 2021-05-19
Final revision date: 2021-10-06
Acceptance date: 2021-10-17
Online publication date: 2021-10-29
Corresponding author
KEYWORDS
TOPICS
ABSTRACT
Introduction:
Kidney tumor is among the 10 most common cancers. Among kidney tumors, renal cell carcinoma (RCC) is one of the most common types, with an alarming increasing incidence rate. Although the disruption of microbiota is an established factor in the progression of intestinal cancers, its role in other types of cancers has been under-studied.
Material and methods:
In this study, the microbiome disruption and the involvement of SNZ (SCHNARCHZAPFEN) and stromalin (SA) genes in the development of kidney cancer have been focused on using a combination of genetic and bioinformatic analysis. The microbiomes of kidney tumor patients were analyzed using various genetic and bioinformatic variations. Genetic and bioinformatic analyses were performed to identify operational taxonomic units (OTUs), SNZ, SA, and annotated species were determined using 41 samples from a population of kidney tumors.
Results:
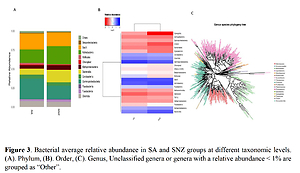
The whole samples from the kidney tumor of patients were screened by PCR amplification and a total of 1317 OTUs were identified. Among them, 379 were common among the two populations, 766 were unique to the SA gene, and 172 to SNZ. SA was more abundant in Gammaproteobacteria and Bacilli, while SNZ had a higher abundance in Bacteroidia and Actinobacteria. Correlation analysis was performed to identify the bacteria that were differentially expressed among the population samples.
Conclusions:
Our study reveals that SA and SNZ are differentially expressed in the microbiome of the kidney tumor, which is associated with the development of kidney tumors such as renal cell carcinoma in human populations.
Kidney tumor is among the 10 most common cancers. Among kidney tumors, renal cell carcinoma (RCC) is one of the most common types, with an alarming increasing incidence rate. Although the disruption of microbiota is an established factor in the progression of intestinal cancers, its role in other types of cancers has been under-studied.
Material and methods:
In this study, the microbiome disruption and the involvement of SNZ (SCHNARCHZAPFEN) and stromalin (SA) genes in the development of kidney cancer have been focused on using a combination of genetic and bioinformatic analysis. The microbiomes of kidney tumor patients were analyzed using various genetic and bioinformatic variations. Genetic and bioinformatic analyses were performed to identify operational taxonomic units (OTUs), SNZ, SA, and annotated species were determined using 41 samples from a population of kidney tumors.
Results:
The whole samples from the kidney tumor of patients were screened by PCR amplification and a total of 1317 OTUs were identified. Among them, 379 were common among the two populations, 766 were unique to the SA gene, and 172 to SNZ. SA was more abundant in Gammaproteobacteria and Bacilli, while SNZ had a higher abundance in Bacteroidia and Actinobacteria. Correlation analysis was performed to identify the bacteria that were differentially expressed among the population samples.
Conclusions:
Our study reveals that SA and SNZ are differentially expressed in the microbiome of the kidney tumor, which is associated with the development of kidney tumors such as renal cell carcinoma in human populations.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.