Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Section Editors
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
ONCOLOGY / CLINICAL RESEARCH
Investigation of potential prognostic biomarkers for colorectal cancer
1
Department of Abdominal Radiotherapy, Clinical Oncology School of Fujian Medical University, Fujian Cancer Hospital, 350011, China
2
Department of Radiation Oncology, Mengchao Hepatobiliary Hospital of Fujian Medical University, China
Submission date: 2022-11-17
Final revision date: 2023-05-16
Acceptance date: 2023-06-03
Online publication date: 2023-07-01
Corresponding author
JuHui Chen
Department of Abdominal Radiotherapy, Fujian Cancer Hospital, Fujian Medical University Cancer Hospital, Fuzhou, China
Department of Abdominal Radiotherapy, Fujian Cancer Hospital, Fujian Medical University Cancer Hospital, Fuzhou, China
KEYWORDS
TOPICS
ABSTRACT
Introduction:
Colorectal cancer (CRC) is the third leading cause of cancer-related death. Since CRC is largely asymptomatic until the alert features develop to an advanced stage, implementation of a screening program is important to reduce cancer morbidity and mortality. Current screening methods have significant limitations.
Material and methods:
CRC-related microarray datasets were collected from the GEO database and differentially expressed genes (DEGs) were identified. Next, Venn analysis, functional enrichment analysis, protein interaction network (PPI) analysis, and survival analysis were performed.
Results:
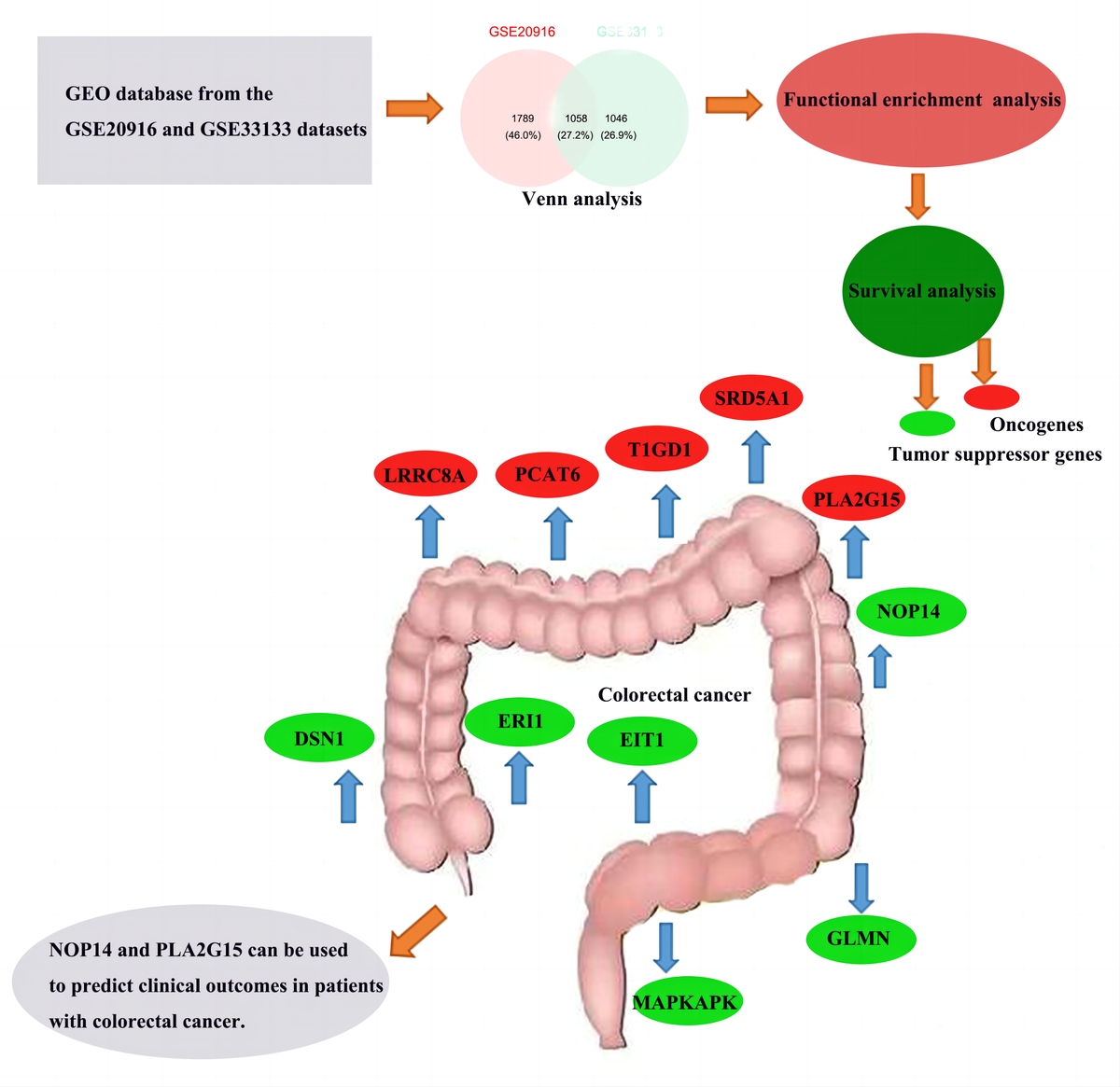
A total of 5267 and 4233 DEGs were identified in two datasets (GSE20916, GSE33133). The intersection of up-regulated genes in the two datasets was obtained by Venn Analysis as 1058 DEGs. Among the 1058 genes, 992 genes with survival and clinical information in TCGA were screened. Eleven DEGs were identified as potential prognostic markers. Model results show that the time period with the most obvious prognostic effect is 5 years, and the AUC value is the highest. ROC curve results are consistent with the model results of the survival analysis. The survival curve showed that LRRC8A, PCAT6, PLA2G15, SRD5A1, T1GD1 may be oncogenes, and DSN1, ERI1, EIT1, GLMN, MAPKAPK, NOP14 may be tumor suppressor genes.
Conclusions:
This study discovers novel prognostic markers through Cox regression and survival analysis, and provides a theoretical basis for the treatment of CRC.
Colorectal cancer (CRC) is the third leading cause of cancer-related death. Since CRC is largely asymptomatic until the alert features develop to an advanced stage, implementation of a screening program is important to reduce cancer morbidity and mortality. Current screening methods have significant limitations.
Material and methods:
CRC-related microarray datasets were collected from the GEO database and differentially expressed genes (DEGs) were identified. Next, Venn analysis, functional enrichment analysis, protein interaction network (PPI) analysis, and survival analysis were performed.
Results:
A total of 5267 and 4233 DEGs were identified in two datasets (GSE20916, GSE33133). The intersection of up-regulated genes in the two datasets was obtained by Venn Analysis as 1058 DEGs. Among the 1058 genes, 992 genes with survival and clinical information in TCGA were screened. Eleven DEGs were identified as potential prognostic markers. Model results show that the time period with the most obvious prognostic effect is 5 years, and the AUC value is the highest. ROC curve results are consistent with the model results of the survival analysis. The survival curve showed that LRRC8A, PCAT6, PLA2G15, SRD5A1, T1GD1 may be oncogenes, and DSN1, ERI1, EIT1, GLMN, MAPKAPK, NOP14 may be tumor suppressor genes.
Conclusions:
This study discovers novel prognostic markers through Cox regression and survival analysis, and provides a theoretical basis for the treatment of CRC.
REFERENCES (53)
1.
Pourhoseingholi MA, Zali MR. Colorectal cancer screening: time for action in Iran. World J Gastrointest Oncol 2012; 4: 82-3.
2.
Marley AR, Nan H. Epidemiology of colorectal cancer. Int J Mol Epidemiol Genet 2016; 7: 105-14.
3.
Voorneveld PW, Jacobs RJ, Kodach LL, et al. A meta-analysis of SMAD4 immunohistochemistry as a prognostic marker in colorectal cancer. Transl Oncol 2015; 8: 18-24.
4.
Zarkavelis G, Boussios S, Papadaki A, et al. Current and future biomarkers in colorectal cancer. Ann Gastroenterol 2017; 30: 613-21.
5.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019; 69: 7-34.
6.
Lad J, Serra S, Quereshy F, et al. Polarimetric biomarkers of peri-tumoral stroma can correlate with 5-year survival in patients with left-sided colorectal cancer. Sci Rep 2022; 12: 12652.
7.
Huang Y, Zhou J, Zhong H, et al. Identification of a novel lipid metabolism-related gene signature for predicting colorectal cancer survival. Front Genet 2022; 13: 989327.
8.
Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg 2009; 22: 191-7.
9.
Liu H, Li Y, Lv Y, et al. LncRNA AK077216 affects the survival of colorectal adenocarcinoma patients via miR-34a. Arab J Gastroenterol 2022; 23: 65-9.
10.
Shussman N, Wexner SD. Colorectal polyps and polyposis syndromes. Gastroenterol Rep 2014; 2: 1-15.
11.
Hibner G, Kimsa-Furdzik M, Francuz T. Relevance of microRNAs as potential diagnostic and prognostic markers in colorectal cancer. Int J Mol Sci 2018; 19: 2944.
12.
Duan L, Yang W, Wang X, et al. Advances in prognostic markers for colorectal cancer. Exp Rev Mol Diagnostics 2019; 19: 313-24.
13.
Carlomagno N, Schonauer F, Tammaro V, et al. A multidisciplinary approach to an unusual medical case of locally advanced gastric cancer: a case report. J Med Case Rep 2015; 9: 13.
14.
Santangelo M, Esposito A, Tammaro V, et al. What indication, morbidity and mortality for central pancreatectomy in oncological surgery? A systematic review. Int J Surg 2016; 28: S172-6.
15.
Issa IA, Noureddine M. Colorectal cancer screening: an updated review of the available options. World J Gastroenterol 2017; 23: 5086.
16.
Das V, Kalita J, Pal M. Predictive and prognostic biomarkers in colorectal cancer: a systematic review of recent advances and challenges. Biomed Pharmacothe 2017; 87: 8-19.
17.
Santangelo M, Romano G, Vescio G, et al. Functional results of colorectal and coloanal anastomosis with and without pouch. Ann Ital Chirur 2001; 72: 443-8.
18.
FDA-NIH Biomarker Working Group. BEST (Biomarkers, endpoints, and other tools) resource [Internet] 2016.
19.
Califf RM. Biomarker definitions and their applications. Exp Biol Med 2018; 243: 213-21.
20.
Hossain MA, Islam SMS, Quinn JMW, et al. Machine learning and bioinformatics models to identify gene expression patterns of ovarian cancer associated with disease progression and mortality. J Biomed Inform 2019; 100: 103313.
21.
Horenblas S, Van Tinteren H. Squamous cell carcinoma of the penis. IV. Prognostic factors of survival: analysis of tumor, nodes and metastasis classification system. J Urol 1994; 151: 1239-43.
22.
Chua DT, Sham JS, Kwong DL, et al. Volumetric analysis of tumor extent in nasopharyngeal carcinoma and correlation with treatment outcome. Int J Radiation Oncol Biol Phys 1997; 39: 711-9.
23.
Rich JT, Neely JG, Paniello RC, et al. A practical guide to understanding Kaplan-Meier curves. Otolaryngol Head Neck Surg 2010; 143: 331-6.
24.
Chen L, Lu D, Sun K, et al. Identification of biomarkers associated with diagnosis and prognosis of colorectal cancer patients based on integrated bioinformatics analysis. Gene 2019; 692: 119-25.
25.
Zheng Y, Cai T, Feng Z. Application of the time‐dependent ROC curves for prognostic accuracy with multiple biomarkers. Biometrics 2006; 62: 279-87.
26.
Barrett T, Wilhite S E, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets – update. Nucleic Acids Res 2012; 41: D991-5.
27.
Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008; 455: 1061-8.
28.
Heberle H, Meirelles GV, da Silva FR, et al. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics 2015; 16: 169.
29.
Young MD, Wakefield MJ, Smyth GK, et al. Gene ontology analysis for RNA-seq: accounting for selection bias Genome Biol 2010; 11: R14.
30.
Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nat Genet 2000; 25: 25-9.
31.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000; 28: 27-30.
32.
Safari-Alighiarloo N, Taghizadeh M, Rezaei-Tavirani M, et al. Protein-protein interaction networks (PPI) and complex diseases. Gastroenterol Hepatol Bed Bench 2014; 7: 17-31.
33.
Mering C, Huynen M, Jaeggi D, et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res 2003; 31: 258-61.
34.
Royston P, Lambert P C. Flexible parametric survival analysis using Stata: beyond the Cox model. College Station, TX: Stata press 2011.
35.
Shahraki H R, Salehi A, Zare N. Survival prognostic factors of male breast cancer in Southern Iran: a LASSO-Cox regression approach. Asian Pacific J Cancer Prev 2015; 16: 6773-7.
36.
Meier L, Van De Geer S, Bühlmann P. The group lasso for logistic regression. J R Statist Soc B 2008; 70: 53-71.
37.
Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394-424.
38.
Dai GP, Wang LP, Wen YU, et al. Identification of key genes for predicting colorectal cancer prognosis by integrated bioinformatics analysis. Oncol Lett 2020; 19: 388-98.
39.
Li J, Fang R, Wang J, Deng L. NOP14 inhibits melanoma proliferation and metastasis by regulating Wnt/beta-catenin signaling pathway. Braz J Med Biol Res 2018; 52: e7952.
40.
Lei JJ, Peng RJ, Kuang BH, et al. NOP14 suppresses breast cancer progression by inhibiting NRIP1/Wnt/beta-catenin pathway. Oncotarget 2015; 6: 25701-14.
41.
Du Y, Liu Z, You L, et al. Pancreatic cancer progression relies upon mutant p53-induced oncogenic signaling mediated by NOP14. Cancer Res 2017; 77: 2661-73.
42.
Lu C, Liao W, Huang Y, et al. Increased expression of NOP14 is associated with improved prognosis due to immune regulation in colorectal cancer. BMC Gastroenterol 2022; 22: 207.
43.
Zhu X, Jia W, Yan Y, et al. NOP14 regulates the growth, migration, and invasion of colorectal cancer cells by modulating the NRIP1/GSK-3/-catenin signaling pathway. Eur J Histochem 2021; 65: 3246.
44.
Navarro-Mendoza MI, Pérez-Arques C, Panchal S, et al. Early diverging fungus Mucor circinelloides lacks centromeric histone CENP-A and displays a mosaic of point and regional centromeres. Curr Biol 2019; 29: 3791-802.
45.
Chuang TP, Wang JY, Jao SW, et al. Over-expression of AURKA, SKA3 and DSN1 contributes to colorectal adenoma to carcinoma progression. Oncotarget 2016; 7: 45803.
46.
Li FN, Zhang QY, Li O, et al. ESRRA promotes gastric cancer development by regulating the CDC25C/CDK1/CyclinB1 pathway via DSN1. Int J Biol Sci 2021; 17: 1909.
47.
Sun C, Huang S, Ju W, et al. Elevated DSN1 expression is associated with poor survival in patients with hepatocellular carcinoma. Human Pathol 2018; 81: 113-20.
48.
Peng Q, Wen T, Liu D, et al. DSN1 is a prognostic biomarker and correlated with clinical characterize in breast cancer. Int Immunopharmacol 2021; 101: 107605.
49.
Murakami M, Sato H, Taketomi Y. Updating phospholipase A2 biology. Biomolecules 2020; 10: 1457.
50.
Shayman JA, Tesmer JJ. Lysosomal phospholipase A2. Biochim Biophys Acta Mol Cell Biol Lipids 2019; 1864: 932-40.
51.
Shayman JA, Tesmer JJG. Lysosomal phospholipase A2. Biochim Biophys Acta Mol Cell Biol Lipids 2019;,1864: 932-40.
52.
Jang JE, Kim HP, Han SW, et al. NFATC3-PLA2G15 fusion transcript identified by RNA sequencing promotes tumor invasion and proliferation in colorectal cancer cell lines. Cancer Res Treat 2019; 51: 391-401.
53.
Dalal N, Jalandra R, Sharma M, et al. Omics technologies for improved diagnosis and treatment of colorectal cancer: Technical advancement and major perspectives. Biomed Pharmacother 2020; 131: 110648.
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.