The 5-year recurrence rate of ischemic stroke (IS) patients is more than 30% in China [1]. Recurrent ischemic stroke (RIS) is associated with a higher risk of mortality and disability. Clopidogrel, an antiplatelet agent that prevents adenosine diphosphate (ADP)-induced platelet aggregation by irreversibly binding to the ADP receptor P2Y12 on the platelet membrane, has always been an important secondary prevention strategy for IS. However, many regular clopidogrel users have the same platelet reactivity as non-treated individuals and they cannot avoid blood clots because the platelet aggregation was not inhibited. This phenomenon is called “high on-treatment platelet reactivity (HPR)” [2], which is an independent risk factor for RIS [3].
Platelet endothelial aggregation receptor 1 (PEAR1) is a membrane protein encoded by the PEAR1 gene and is expressed on the surface of platelets and endothelial cells; PEAR1 rs12041331 is one of the most strongly expressed PEAR1 variants at many reported loci. Polymorphism of PEAR1 rs12041331 is believed to contribute to HPR in patients with ischemic stroke; however, this is controversial [4, 5]. One potential reason for this divergence may be that only about 12% of HPR occurrences are associated with gene polymorphism [6]. PEAR1 rs12041331 is the first functional CpG-SNP related to platelet function whose regulatory mechanism depends on DNA methylation [7]. The major G allele forming the CpG pair is associated with higher gene expression in platelets in comparison to the minor A allele [7]. We speculate that PEAR1 rs12041331 polymorphisms and DNA methylation are associated with the risk of HPR and recurrent stroke. As far as we know, there is no research focusing on the potential association between PEAR1 DNA methylation and clopidogrel HPR in ischemic stroke patients.
Our study is the first to investigate the relationship between PEAR1 rs12041331, locus-specific DNA methylation, and gene expression alterations in RIS patients, and its possible correlation with clopidogrel HPR.
Methods
A total of 90 patients diagnosed with acute minor ischemic stroke were recruited and sorted into two groups: 46 cases of primary ischemic stroke (PIS group) and 44 cases of RIS. The RIS group consisted of patients either with a history of IS and relapsing or with newly diagnosed IS after onset of the first infarction [8]. Patients did not have hematologic disorders and were not taking any other antiplatelet or anticoagulant drug. All patients were placed on clopidogrel 75 mg once daily for at least 8 days and atorvastatin calcium 20 mg daily after admission. Evaluation of ADP-triggered platelet aggregation inhibition rate was performed through thromboelastography on the eighth day after admission. The HPR and normal on-treatment platelet reactivity (NPR) were defined using the 5 μmol/l ADP-induced platelet inhibition rate as the criterion (< 50% and ≥ 50%, respectively).
On the eighth day after admission whole blood samples were taken and DNA was extracted from them using a TIANamp Genomic DNA Kit (Tiangen, DP304-03, Beijing) according to the manufacturer’s instructions. The genotype of the PEAR1 rs12041331 polymorphism was confirmed by polymerase chain reaction (PCR) amplification and Sanger sequencing according to the manufacturer’s instructions. Primer 5.0 was used to design primers: PEAR1-F (5′ GAGGAGTGGGACTGAGAAT 3′) and PEAR1-R (5′ GACAGGCAGGGTGTTGAG 3′).
CpG sites were analyzed by pyrosequencing. Bisulfite conversion reaction was performed using an EpiTect Bisulfite Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Converted DNA was amplified by PCR using the PyroMark PCR Kit (Qiagen). PCR primers were as follows: forward: 5′ GTGGGGGGGATTAGAGTT 3′; reverse: 5′ AACCCCTAAAAAAATCCCTTCTACTATCTC 3′. The cycling conditions were recommended by the manufacturer. 20 μl of PCR product was pyrosequenced using the PyroMark Gold Q96 Kit and a pyrosequencer (QIAGEN).
Ribonucleic acid (RNA) was extracted from serum samples using miRNeasy Serum/Plasma Kits (QIAGEN, Hilden, Germany, catalog number 217184) according to the manufacturer’s instructions. Quantitative PCR (qPCR) was conducted using the PerfectStart Green qPCR SuperMix in a Light Cycler 480 Gene Scanning system (Roche, Swiss). Primers used were as follows: forward: 5′ CCAGGATGGGAGCTGTAT 3′, and reverse: 5′GGAGCAGTTAGCACCAAAT 3′. Glyceraldehyde-phosphate dehydrogenase (GAPDH) was used as an internal reference (forward: 5′CCTCACAGTTGCCATGTAGA3′, and reverse: 5′TGGTACATGACAAGGTGCG 3′). Real-time PCR reactions were prepared, and the qPCR conditions were according to the manufacturer’s recommendations. Fluorescence intensity was then detected with a melting curve from 60°C to 97°C.
Ethics statement
The studies involving human participants were reviewed and approved by the Ethics Committee of ZhuZhou Central Hospital Affiliated to Central South University Xiangya School of Medicine. The study participants provided their written informed consent to be included in this study.
Statistical analysis
The PEAR1 rs12041331 genotypic distribution was in Hardy-Weinberg equilibrium. Differences in the distributions of demographic characteristics, frequencies of genotypes and levels of DNA methylation mRNA between two independent groups were evaluated using the independent t-test or Mann-Whitney U test (for continuous variables) and the χ2 test or Fisher’s exact test (for categorical variables). Statistical analysis was performed using IBM SPSS software (SPSS 23.0 for Windows; IBM, Chicago, IL, USA), and p ≤ 0.05 was considered statistically significant.
Results
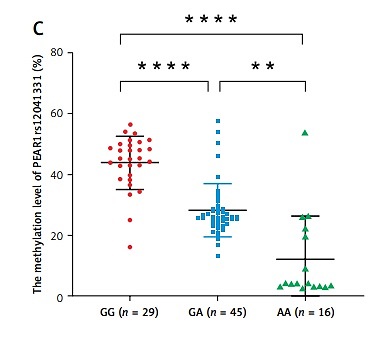
Age (p = 0.019) and the proportion of HPR (p = 0.029) were higher in the RIS group. There were no differences between the PIS and RIS groups in other traditional risk factors for AIS, including gender, body mass index (BMI), hypertension, diabetes mellitus, smoking history, high-density lipoprotein cholesterol (HDL-c), and low-density lipoprotein cholesterol (LDL-c) (p > 0.05) (Table I). A significant fraction of HPR (47.8%, p = 0.014) patients were homozygous for the major G allele (GG) of PEAR1 rs12041331; this allele has been related to HPR (p = 0.007) (Table I, Figure 1 A). The PEAR1 DNA methylation level (33.53 ±14.28% vs. 26.11 ±14.53%, p = 0.044) and mRNA expression (6.28 ±1.11 vs. 5.75 ±1.36, p = 0.046) were significantly higher in the HPR group compared to the NPR group (Figure 1 B). The PEAR1 DNA methylation level was the highest for GG homozygotes, who had significantly higher methylation levels than GA and AA individuals (Figure 1 C). The PEAR1 mRNA expression was independent of PEAR1 rs12041331 genotype (Figure 1 D) and DNA methylation (r = 0.157, p = 0.139). There was no significant difference when comparing the PEAR1 rs12041331 genotypes, methylation and mRNA levels between the PIS and RIS groups (data not shown). Multivariate logistic regression analysis showed that carriage of the G allele, higher mRNA expression, and RIS are independently associated with a higher risk of HPR (Figure 1 E).
Table I
Baseline characteristics of patients within the different groups
[i] Values are presented as mean ± SD or number of patients (percentage) as appropriate. P-values represent the statistical difference of each variable by PIS group and RIS group or HPR group and NPR group. SNP – single nucleotide polymorphism, BMI – body mass index, HDL-C – high-density lipoprotein cholesterol, LDL-C – low-density lipoprotein cholesterol, ADP – adenosine diphosphate, HbA1c – glycated hemoglobin, PIS – primary ischemic stroke group, RIS – recurrent ischemic stroke, HPR – high on-treatment platelet reactivity, NPR – normal on-treatment platelet reactivity.
Figure 1
A – A higher proportion of HPR patients were homozygous for the major G allele (GG) of PEAR1 rs12041331 (p = 0.014). The difference in genotype between the RIS and PIS groups was not statistically significant (p = 0.247). B – The PEAR1 DNA methylation level (p = 0.044) and mRNA expression (p = 0.046) were higher in the HPR group compared to the NPR group. C – The PEAR1 DNA methylation level is highest for GG homozygotes, who had significantly higher methylation levels than GA and AA individuals (p < 0.001). D – The mRNA expression was independent of PEAR1 rs12041331 genotype. E – Forest plot of associations between HPR and common conditions
Discussion
In this study we were the first to analyze the PEAR1 rs12041331 genetic polymorphism, DNA methylation, and mRNA expression level and its correlation with HPR in patients with RIS. Our data indicate that the G allele is associated with a higher prevalence of HPR. Li et al. [5] also observed an association between the GG genotype and HPR in patients with IS treated with aspirin. It is worth noting that patients with coronary artery disease carrying the A allele associated with lower platelet reactivity are at high risk for cardiovascular events [5]. This high risk was also reported in a retrospective study showing that the A allele was more frequent than the G allele in RIS patients [9]. This result contradicts the idea that HPR is linked to a poorer clinical outcome including ischemic stroke. One possible explanation is that the A allele could positively affect the migration of endothelial cells and promote thrombosis [10]. In our study, we did not observe an association between the PEAR1 rs12041331 polymorphism and RIS, although we did find that the G allele is related to HPR linked with RIS. As suggested in two other studies published by Yue et al., PEAR1 rs12041331 AA carriers with stroke are highly sensitive to aspirin therapy [5]. Another study proposed that genetic variations in PEAR1 rs12041331 do not contribute to atherothrombotic risk in stroke patients [11]. Unlike our study, the latter did not involve measurements of platelet reactivity; none of these studies considered DNA methylation and mRNA levels. Nevertheless, scant research has been devoted to the role of PEAR1 on the risk of RIS. Contrary to our study, Du et al. found that the polymorphism at PEAR1 was independently associated with RIS in Chinese patients. Therefore, we speculate that the antiplatelet effect is influenced not only by the mutation of PEAR1, but also by DNA methylation [12].
Consistent with our findings, a study showed that a low methylation level was significantly associated with the minor A allele [7]. However, inconsistent with an vitro experiment showing greater platelet expression of the PEAR1 protein in relation to the G allele of the PEAR1 rs12041331 [13], no direct correlation was found between the G allele, hypermethylation and mRNA levels in our study. One likely reason for this inconsistency is the complex genetic regulation of gene expression by other loci or epigenetic regulatory mechanisms. Despite this, we speculate that there is a correlation between methylation and mRNA levels of PEAR1 because the hypermethylation and high mRNA levels are both associated with HPR in our results. However, this hypothesis differs from the generally accepted theory that gene expression is negatively associated with DNA methylation. To explain this phenomenon, Izzi et al. proposed that the methylation level of PEAR1 is positively correlated with its expression during the early stages of cell differentiation [14]. Regrettably, no association between RIS, genotype and methylation of PEAR1 was observed in our study, possibly because RIS is often caused by multiple factors, and a single genetic factor may not be sufficient to detect the risk associated with RIS.
Our study has some limitations such as an insufficient sample size and the fact that we studied only one locus (PEAR1 rs12041331). Further studies should focus on protein levels as well. In conclusion, carriers of the G allele of PEAR1 rs12041331 and promoter hypermethylation may also have increased HPR risk. However, the association between PEAR 1 rs12041331 and RIS has not been proved.