Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Section Editors
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
CLINICAL RESEARCH
Identifying core genes in sepsis by LASSO regression and SVM-RFE algorithm
1
Jiangxi Provincial People’s Hospital, The First Affiliated Hospital of Nanchang Medical College, Nanchang, Jiangxi, China
Submission date: 2023-09-15
Final revision date: 2024-01-04
Acceptance date: 2024-03-25
Online publication date: 2024-12-13
Corresponding author
Jie Yu
Jiangxi Provincial People’s Hospital The First Affiliated Hospital of Nanchang Medical College Nanchang Jiangxi, China
Jiangxi Provincial People’s Hospital The First Affiliated Hospital of Nanchang Medical College Nanchang Jiangxi, China
KEYWORDS
sepsisLASSO regressionsupport vector machine recursive feature elimination (SVM-RFE) algorithmbioinformatics analysisfunctional analysis
TOPICS
ABSTRACT
Introduction:
Sepsis is a major disease in intensive care units (ICU), with high morbidity and mortality. However, the core genes associated with the sepsis diagnosis remain unclear.
Material and methods:
By merging five datasets, gene expression profiles were obtained: GSE28750, GSE57065, GSE64457, GSE65682 and GSE95233. Differentially expressed genes (DEGs) were identified using the Limma package in R. To examine the enriched functions, both Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) were employed. Subsequently, the protein-protein interaction network (PPI) was constructed, and module analysis was carried out using STRING and Cytoscape. Furthermore, core genes were identified using support vector machine recursive feature elimination (SVM-RFE) analysis and the least absolute shrinkage and selection operator (LASSO) model. To verify the diagnostic significance of these essential genes, we conducted an analysis of the receiver operating characteristic curve (ROC).
Results:
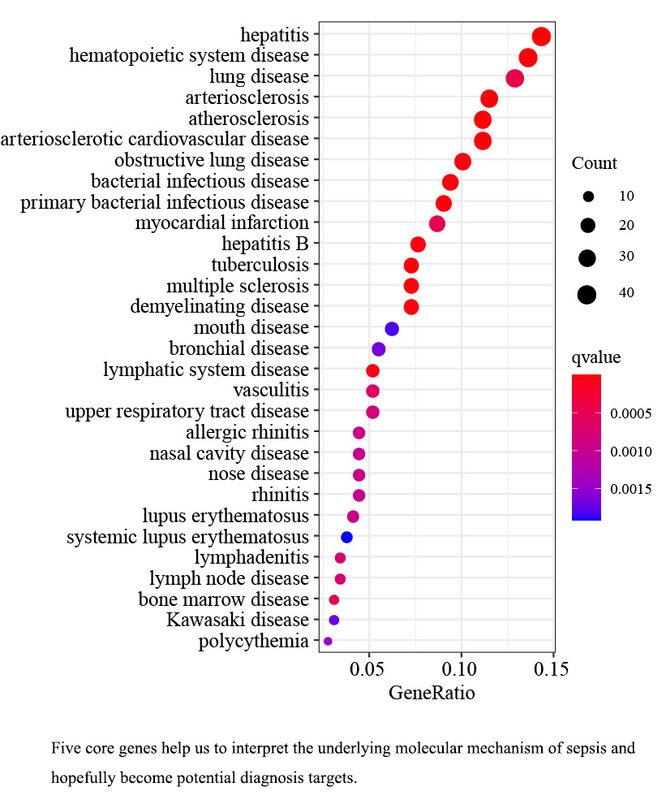
We analyzed 230 DEGs, consisting of 183 upregulated DEGs and 47 downregulated DEGs. The GO and KEGG analyses revealed that the DEGs were enriched in immune-related pathways and functions. The DEGs formed a PPI network consisting of 180 protein nodes and 351 interaction edges. Ultimately, we identified the five critical core genes (C3AR1, CHPT1, RAB32, SLC22A4, and SRPK1) common between both algorithms. The analysis of the ROC curve demonstrated that the AUC values for the five fundamental genes were as follows: 0.881, 0.876, 0.946, 0.927, and 0.931, respectively.
Conclusions:
The five core genes screened in this study will help us to interpret the underlying molecular mechanism of sepsis and hopefully become potential diagnostic targets.
Sepsis is a major disease in intensive care units (ICU), with high morbidity and mortality. However, the core genes associated with the sepsis diagnosis remain unclear.
Material and methods:
By merging five datasets, gene expression profiles were obtained: GSE28750, GSE57065, GSE64457, GSE65682 and GSE95233. Differentially expressed genes (DEGs) were identified using the Limma package in R. To examine the enriched functions, both Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) were employed. Subsequently, the protein-protein interaction network (PPI) was constructed, and module analysis was carried out using STRING and Cytoscape. Furthermore, core genes were identified using support vector machine recursive feature elimination (SVM-RFE) analysis and the least absolute shrinkage and selection operator (LASSO) model. To verify the diagnostic significance of these essential genes, we conducted an analysis of the receiver operating characteristic curve (ROC).
Results:
We analyzed 230 DEGs, consisting of 183 upregulated DEGs and 47 downregulated DEGs. The GO and KEGG analyses revealed that the DEGs were enriched in immune-related pathways and functions. The DEGs formed a PPI network consisting of 180 protein nodes and 351 interaction edges. Ultimately, we identified the five critical core genes (C3AR1, CHPT1, RAB32, SLC22A4, and SRPK1) common between both algorithms. The analysis of the ROC curve demonstrated that the AUC values for the five fundamental genes were as follows: 0.881, 0.876, 0.946, 0.927, and 0.931, respectively.
Conclusions:
The five core genes screened in this study will help us to interpret the underlying molecular mechanism of sepsis and hopefully become potential diagnostic targets.
REFERENCES (32)
1.
Rhodes A, Evans LE, Alhazzani W, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Intensive Care Med 2017; 43: 304-77.
2.
Vincent JL, Opal SM, Marshall JC, Tracey KJ. Sepsis definitions: time for change. Lancet 2013; 381: 774-5.
3.
Scicluna BP, van Vught LA, Zwinderman AH, et al. Classification of patients with sepsis according to blood genomic endotype: a prospective cohort study. Lancet Respir Med 2017; 5: 816-26.
4.
Pettilä V, Hynninen M, Takkunen O, Kuusela P, Valtonen M. Predictive value of procalcitonin and interleukin 6 in critically ill patients with suspected sepsis. Intensive Care Med 2002; 28: 1220-5.
5.
Hu Q, Gong W, Gu J, et al. Plasma microRNA profiles as a potential biomarker in differentiating adult-onset Still’s disease from sepsis. Front Immunol 2018; 9: 3099.
6.
Sinha M, Jupe J, Mack H, Coleman TP, Lawrence SM, Fraley SI. Emerging technologies for molecular diagnosis of sepsis. Clin Microbiol Rev 2018; 31: e00089-17.
7.
Schenz J, Weigand MA, Uhle F. Molecular and biomarker-based diagnostics in early sepsis: current challenges and future perspectives. Exp Rev Mol Diagn 2019; 19: 1069-78.
8.
Nie MW, Han YC, Shen ZJ, Xie HZ. Identification of circRNA and mRNA expression profiles and functional networks of vascular tissue in lipopolysaccharide-induced sepsis. J Cell Mol Med 2020; 24: 7915-27.
9.
Goujon M, McWilliam H, Li W, et al. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 2010; 38: W695-699.
10.
Lee J, Banerjee D. Metabolomics and the microbiome as biomarkers in sepsis. Crit Care Clin 2020; 36: 105-13.
11.
Qin Y, Guo X, Yu Y, et al. Screening key genes and microRNAs in sepsis by RNA-sequencing. J Chin Med Assoc 2020; 83: 41-7.
12.
Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019; 47: D607-13.
13.
Li J, Liu C, Chen Y, et al. Tumor Characterization in breast cancer identifies immune-relevant gene signatures associated with prognosis. Front Genet 2019; 10: 1119.
14.
Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010; 33: 1-22.
15.
Lao J, Chen Y, Li ZC. A deep learning-based radiomics model for prediction of survival in glioblastoma multiforme. Sci Rep 2017; 7: 10353.
16.
Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 1999; 286: 531-7.
17.
Huang S, Cai N, Pacheco PP, Narrandes S, Wang Y, Xu W. Applications of Support Vector Machine (SVM) learning in cancer genomics. Cancer Genomics Proteomics 2018; 15: 41-51.
18.
Poovizhi S, Ganesh Babu Tr. An efficient skin cancer diagnostic system using bendlet transform and support vector machine. An Acad Brasil Cienc 2020; 92: e20190554.
19.
Zhang Y, Wu Y, Gong ZY, et al. Distinguishing rectal cancer from colon cancer based on the support vector machine method and RNA-sequencing data. Curr Med Sci 2021; 41: 368-74.
20.
Jiang Y, Xie J, Han Z, et al. Immunomarker support vector machine classifier for prediction of gastric cancer survival and adjuvant chemotherapeutic benefit. Clin Cancer Res 2018; 24: 5574-84.
21.
Li M, Huang H, Ke C, et al. Identification of a novel four-gene diagnostic signature for patients with sepsis by integrating weighted gene co-expression network analysis and support vector machine algorithm. Hereditas 2022; 159: 14.
22.
Srisawat N, Kulvichit W, Tungsanga S, et al. The role of neutrophil chemotaxis activity as an immunologic biomarker to predict mortality in critically-ill patients with severe sepsis. J Crit Care 2020; 56: 215-21.
23.
Saito M, Inoue S, Yamashita K, Kakeji Y, Fukumoto T, Kotani J. IL-15 improves aging-induced persistent T cell exhaustion in mouse models of repeated sepsis. Shock (Augusta, Ga) 2020; 53: 228-35.
24.
Chen L, Ke H, Zhang Y, et al. Orai1 overexpression improves sepsis-induced T-lymphocyte immunosuppression and acute organ dysfunction in mice. Heliyon 2022; 8: e12082.
25.
Lu G, Li Q, Liu J, Jia Y, Tang J, Zhang X. Inhibition of endoplasmic reticulum stress and the downstream pathways protects CD4(+) T cells against apoptosis and immune dysregulation in sepsis. IUBMB Life 2022; 74: 1070-80.
26.
Chen CW, Xue M, Zhang W, Xie J, Coopersmith CM, Ford ML. 2B4 but not PD-1 blockade improves mortality in septic animals with preexisting malignancy. JCI Insight 2019; 4: e127867.
27.
Brennan FH, Jogia T, Gillespie ER, et al. Complement receptor C3aR1 controls neutrophil mobilization following spinal cord injury through physiological antagonism of CXCR2. JCI Insight 2019; 4: e98254.
28.
Cadenas C, Vosbeck S, Hein EM, et al. Glycerophospholipid profile in oncogene-induced senescence. Biochim Biophys Acta 2012; 1821: 1256-68.
29.
Li Q, Lei F, Tang Y, et al. Megalin mediates plasma membrane to mitochondria cross-talk and regulates mitochondrial metabolism. Cell Mol Life Sci 2018; 75: 4021-40.
30.
Nigam SK. The SLC22 transporter family: a paradigm for the impact of drug transporters on metabolic pathways, signaling, and disease. Ann Rev Pharmacol Toxicol 2018; 58: 663-87.
31.
Gottier Nwafor J, Nowik M, Anzai N, Endou H, Wagner CA. Metabolic acidosis alters expression of Slc22 transporters in mouse kidney. Kidney Blood Pressure Res 2020; 45: 263-74.
32.
Guo W, Hu Z. SRPK1 promotes sepsis-induced acute lung injury via regulating PI3K/AKT/FOXO3 signaling. Immunopharmacol Immunotoxicol 2023; 45: 203-12.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.