Introduction
Coronary artery disease (CAD) is a pathological process characterized by obstructive or non-obstructive atherosclerotic plaque accumulation in the epicardial arteries. The disease can have long, stable periods but can also become unstable at any time, typically due to an acute atherothrombotic event caused by plaque rupture or erosion, which can present clinically as either chronic coronary syndrome (CCS) or acute coronary syndrome (ACS) [1]. Acute coronary syndrome is a medical emergency requiring immediate intervention; CCS also needs to be taken seriously since CAD is often progressive even in apparently clinically silent periods.
In the development and progression of CAD, emerging evidence suggests that exosomes have played crucial roles [2]. Exosomes are small membrane microvesicles (30–120 nm), produced by almost all cell types, that deliver their cargoes (proteins, DNA, and non-coding RNAs) to be involved in intercellular communication [3]. Recent studies have shown that exosomes might play important roles in cardiovascular diseases, and exosomal cargoes emerged as biomarkers or for use in therapeutic approaches [4–6]. Exosomal miRNAs have been reported to be potential biomarkers in ACS [7]; and exosomal lncRNAs may be involved in atherosclerosis progression [8–10], which is the key prerequisite for CCS. Several studies have investigated the role of lncRNAs serving as biomarkers to diagnose CAD [11–14], however, scarce research has been performed on the role of exosomal lncRNAs in CCS patients.
In the present study, we compared the expression of circulating exosomal lncRNAs in CCS patients and control subjects, and investigated the relationship between exosomal lncRNAs and the clinical parameters in CCS patients. Our findings indicate a potential role of circulating exosomal lncRNAs in CCS.
Material and methods
Study subjects
This is a case-control study. All CCS cases and control subjects were consecutively enrolled from Beijing Chao-Yang Hospital (Capital Medical University, Beijing, China) between January and December 2017. All the participants in this investigation had been informed and had given written consent. The study was conducted in accordance with the Declaration of Helsinki, and the research protocol was approved by the Ethics Committee of Beijing Chao-Yang Hospital (Capital Medical University, Beijing, China).
Chronic coronary syndrome was defined according to the European Society of Cardiology (ESC) Clinical Practice Guidelines in 2019 [1]. Six subcategories of CCS were included: suspected CAD with stable anginal symptoms and or dyspnea (anginal equivalent); new onset heart failure or left ventricular dysfunction and suspected coronary artery disease; asymptomatic and symptomatic patients with stabilized symptoms occurring less than 1 year after an acute coronary syndrome or patients with recent revascularization; asymptomatic and symptomatic patient more than 1 year after initial diagnosis or revascularization; patients with angina and suspected microvascular disease or vasospastic angina; and asymptomatic persons in whom CAD was detected at screening. All CCS patients included in the present study were those with coronary vessel stenosis ≥ 50% on coronarography. The control subjects were non-coronary chest pain patients (NCCP), with chest pain, normal cardiac biomarkers and most importantly, no coronary stenosis as confirmed by coronarography.
The following exclusion criteria were applied: malignant diseases, severe organ failure, thyroid dysfunction, autoimmune diseases, and various acute or chronic infectious diseases, and pregnancy.
A total of 218 participants (137 males and 81 females) were included in the present study, and all the study subjects were divided into three groups: a sequencing group (subjects’ blood samples for exosomal RNA sequencing), including 15 CCS and 15 controls (for sequencing profiles, the blood samples of every five subjects were pooled into one collective sample, and thus “3 CCS” and “3 control” samples were profiled); the first validation group (samples for exosomal RNA validation were taken for the first time), including 20 CCS and 20 controls; the second validation group (samples for the second validation were collected), including 100 CCS and 48 controls.
Exosome experiments
Exosomal isolation and identification were performed. Exosomal isolation was done following the instructions of the commercial kit used (Qiagen Inc., Dusseldorf, Germany). Isolated exosomes were identified by transmission electron microscopy (TEM), nanoparticle tracking analysis (NTA), and Western blotting (WB) of the exosomal marker protein CD63 (Abcam, Cambridge, UK) and HSP70 (Abcam, Cambridge, UK) as described previously [15–17].
Exosomal RNA experiments
Exosomal RNA experiments included exosomal RNA extraction, sequencing, and validation by quantitative RT-PCR (qRT-PCR). Exosomal RNA extraction was carried out according to the protocol of the commercial kit utilized (Qiagen Inc., Dusseldorf, Germany). RNA sequencing was done as described in the methods section of our previous study [18]. The relative expression levels of lncRNA were quantified using ViiA 7 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) based on standard methods. The forward and reverse primers used are listed in Table I (the lncRNA IDs were searched in Ensembl Human GRCh37.p13).
Table I
Forward and reverse primers
Laboratory measurements, echocardiography, and coronary artery angiography
Peripheral venous blood samples were collected after overnight fasting in the first 24 h after admission before coronary artery angiography (CAG) to measure a range of parameters, including lipids, glucose, and creatinine. Blood samples with a volume of 8 ml (collected into EDTA-containing tubes) were retained to obtain plasma for exosome isolation, and were then stored at –80°C until further use. Echocardiography was performed to measure routine parameters, such as the left ventricular end diastolic diameter (LVEDD), left ventricular ejection fraction (LVEF), etc. CAG was performed within 72 h after admission in both CCS patients and control subjects.
Statistical analysis
Statistical analysis was performed using SPSS 24.0 software (IBM, Armonk, NY, USA). All statistical analyses were two-tailed, and p-values < 0.05 were considered statistically significant. Continuous variables with normal distribution were expressed as mean ± standard deviation (SD), and compared by Student’s t-test, whereas those with non-normal distribution were expressed as quartiles and compared by the Mann-Whitney U-test. Categorical variables were expressed as percentages and numbers, and compared using the χ2 test. Spearman correlation was used to calculate the correlation between exosomal lncRNAs and clinical parameters. Receiver operating characteristic curves and areas under the curve (AUCs) were computed to estimate the effectiveness of exosomal lncRNAs for prediction.
Results
Sequence profiling of circulating exosomal lncRNA
The present study included 15 CCS patients and 15 control subjects, whose clinical characteristics are summarized in Table II. No significant differences were detected in the subject characteristics between the two groups; they were well balanced with regard to the main clinical and laboratory characteristics.
Table II
Clinical characteristics of sequencing samples
Before sequence profiling of circulating exosomal lncRNA, exosomes from the plasma were identified by TEM analyses and NTA and WB experiments (Supplementary Figure S1). TEM showed that the exosomes had a spherical shape with a bilayer membrane vesicle structure. The diameter of each of the exosome established by NTA was approximately 100 nm. The results of WB experiments suggested that specific proteins, CD63 and HSP70, were positively expressed in the exosomes in both groups.
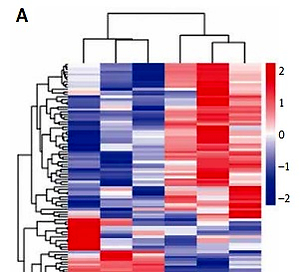
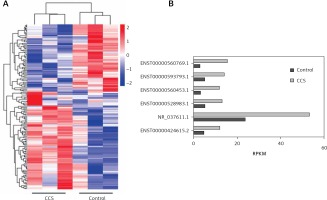
By sequence profiling, we detected a total number of 152 significantly differentially expressed lncRNAs (p < 0.05; fold change > 2) in the plasma exosomes of the two groups, including 90 upregulated and 62 downregulated. Of the 90 upregulated lncRNA identified, we selected six highly expressed lncRNAs with the top fold change (lncRNA ENST00000424-615.2, lncRNA NR_037611.1, lncRNA ENST0000052-8983.1, lncRNA ENST00000560453.1, lncRNA ENST00-000593793.1, and lncRNA ENST0000056-0769.1) to perform the subsequent first validation (Figure 1).
First validation of circulating exosomal lncRNA
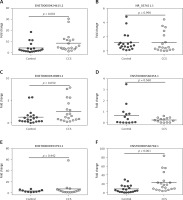
The first validation was performed in 20 CCS patients and 20 control subjects, whose clinical characteristics are presented in Table III. The results of the first validation of the six selected circulating exosomal lncRNAs (ENST00000424615.2, NR_037611.1, ENST00000528983.1, ENST0000056-0453.1, ENST00000593793.1, and ENST00000560769.1) showed that lncRNA ENST00000424615.2 (p < 0.001, fold change = 2.49) and lncRNA ENST00000560769.1 (p < 0.001, fold change = 3.17) were significantly upregulated in AMI patients in the first validation. On the other hand, lncRNA NR_037611.1 (p = 0.990, fold change = 1.67), lncRNA ENST00000528983.1 (p = 0.050, fold change = 1.99), lncRNA ENST00000560453.1 (p = 0.560, fold change = 4.50), and lncRNA ENST-00000593793.1 (p = 0.442, fold change = 1.29) were not (Figure 2). Therefore, the circulating exosomal lncRNAs ENST00000424615.2 and ENST000005-60769.1 were selected for the second validation.
Table III
Clinical characteristics of subjects in the first and second validations
[i] CCS – chronic coronary syndrome, ESR – erythrocyte sedimentation rate, AST – aspartate aminotransferase, ALT – alanine aminotransferase, HDL-C – high-density lipoprotein cholesterol, LDL-C – low-density lipoprotein cholesterol, HbA1c – glycosylated hemoglobin, BUN – blood urea nitrogen, sTSH – thyroid stimulating hormone, CKMB – creatine kinase MB, ACEI/ARB – angiotensin-converting enzyme inhibitors/angiotensin receptor blockers, CCB – calcium antagonist, LVEDD – left ventricular end diastolic diameter, LVEF – left ventricular ejection fraction, NT-BNP – N-terminal proB-type natriuretic peptide.
Second validation of circulating exosomal lncRNA
The second validation was performed in 100 CCS patients and 48 controls, whose clinical characteristics are displayed in Table III.
In the second validation, circulating exosomal lncRNA ENST00000424615.2 (p = 0.063, fold change = 2.39) and lncRNA ENST00000560769.1 (p = 0.003, fold change = 3.07) were both significantly upregulated in CCS patients compared with control subjects (Figure 3).
Figure 3
Circulating exosomal lncRNAs ENST00000424615.2 and ENST00000560769.1, selected in the second validation. A – qRT-PCR analysis of expression of ENST00000424615.2 and ENST00000560769.1. B – Receiver operating characteristic (ROC) curves analyses of ENST00000424615.2 and ENST00000560769.1 for chronic coronary syndrome
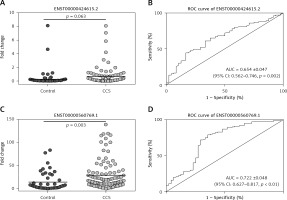
ROC curve analysis showed, for circulating exosomal lncRNA ENST00000424615.2, AUC = 0.654 ±0.047 (95% CI: 0.562–0.746, p = 0.002); for lncRNA ENST00000560769.1, AUC = 0.722 ±0.048 (95% CI: 0.627–0.817, p < 0.001) (Figure 3).
Association between circulating exosomal lncRNA and clinical parameters
In the second validation, circulating exosomal lncRNAs ENST00000424615.2 and ENST-00000560769.1 showed significant relationships with certain parameters in CCS patients. More specifically, exosomal lncRNA ENST00000424615.2 was positively associated with diastolic blood pressure (r = 0.191, p = 0.020), and negatively associated with alanine aminotransferase (ALT) (r = –0.224, p = 0.028) (Table IV). Exosomal lncRNA ENST00000560769.1 was significantly lower in the current drinker group (p = 0.042), but higher in the CCS patients with more diseased vessels (p = 0.028) (Table V).
Table IV
Associations of circulating exosomal lncRNA ENST00000424615.2 and ENST00000560769.1 with clinical parameters of chronic coronary syndrome patients in the second validation
[i] LVEDD – left ventricular end diastolic diameter, LVEF – left ventricular ejection fraction, ESR – erythrocyte sedimentation rate, AST – aspartate aminotransferase, ALT – alanine aminotransferase, HDL-C – high-density lipoprotein cholesterol, LDL-C – low-density lipoprotein cholesterol, HbA1c – glycosylated hemoglobin, BUN – blood urea nitrogen, sTSH – thyroid stimulating hormone, NT-BNP – N-terminal proB-type natriuretic peptide.
Table V
Comparison of expression of circulating exosomal lncRNA ENST00000424615.2 and ENST00000560769.1 between different groups of clinical parameters in chronic coronary syndrome patients in the second validation
Discussion
In the present study, we obtained interesting findings indicating that circulating exosomal lncRNAs ENST00000424615.2 and ENST-00000560769.1 might act as potential biomarkers for CCS. Furthermore, exosomal lncRNA ENST00000560769.1 might be associated with CCS severity, as it was significantly higher in the CCS patients with more diseased vessels.
Increasing evidence has revealed that noncoding RNAs participate in various physiological and pathological processes, playing important roles in human diseases [19–21]. LncRNAs are noncoding RNAs longer than 200 nucleotides, which regulate gene expression at the transcriptional, post-transcriptional, and translational levels. Previous studies have proved that lncRNAs were involved in regulating pathophysiological conditions, such as cancer [22–24], autoimmune diseases [25], and others [26]. In CAD, lncRNAs were found to be differently expressed in atherosclerotic coronary artery plaques [27], and were proved to serve as biomarkers to diagnose CAD [11–14]. Several studies also reported that lncRNAs might be used for the prediction of ACS outcomes [28].
The lncRNA packaged in the exosomes are highly stable due to their lipid bilayers that protect them from enzymatic degradation by RNA enzymes in the bodily fluids. Thus, due to that high stability, exosomal lncRNAs are more suitable to be biomarkers. Previous studies showed that exosomal lncRNAs were potential biomarkers for various diseases [29–31]. In CAD, exosomal lncRNAs were reported to be involved in atherosclerosis progression [8–10], and they might serve as a valuable therapeutic tool for neovascularization [32].
Here, through sequencing profiles and double qRT-PCR validations, we established that circulating exosomal lncRNAs ENST00000424615.2 and ENST00000560769.1 were significantly upregulated in CCS patients. ENST00000424615.2, also named as RP11-38L15.2-001 (Ensembl Human GRCh37. p13), is a lincRNA, and a product of the gene ENSG00000229227.4, located in chromosome 10: 46,937,463-46,947,262 reverse strand. (Ensembl Human GRCh37. p13). ENST00000560769.1, also named as CTD-2033D15.1-001 (Ensembl Human GRCh37. p13), is an antisense lncRNA, and a product of the gene ENSG00000259279.1, located in chromosome 15: 39,885,781-39,886,432 reverse strand (Ensembl Human GRCh37. p13). These two lncRNAs are novel transcripts, on which no relevant research has been performed. In the present study, we found that circulating exosomal lncRNAs ENST00000424615.2 and ENST00000560769.1 were elevated in patients with CCS, and might serve as potential biomarkers for CCS. Moreover, exosomal lncRNA ENST00000560769.1 was significantly higher in the CCS patients with more diseased vessels, and might be associated with a poor prognosis.
Our results suggested that circulating exosomal lncRNAs may serve as potential biomarkers for CCS and even be associated with the severity of CCS. However, the present study also has some limitations: this study was a clinical observation, the mechanism by which circulating exosomal lncRNAs regulate CCS was not elucidated, and further research is needed to clarify these regulatory mechanisms; this study had a small sample size and lacked follow-up after discharge; thus the relationship between exosomal lncRNAs and prognosis was not evaluated, and a further large sample size study with long-term follow-up should be conducted.
In conclusion, our data showed that circulating exosomal lncRNAs ENST00000424615.2 and ENST00000560769.1 were elevated in patients with CCS, and might serve as potential biomarkers for CCS. Moreover, exosomal lncRNA ENST00000560769.1 was significantly higher in the CCS patients with more diseased vessels, and might be associated with a poor prognosis.