Introduction
Lung cancer accounts for over 27% of all cancer cases and remains a health issue all over the world [1]. Fortunately, many diagnostic methods, such as X-ray, bronchoscopy, radionuclide, sputum cytology, and mediastinoscopy, have improved the diagnosis of lung cancer for patients [2, 3]. Rehabilitation massage, as a non-invasive intervention, improves the quality of life in lung cancer patients by alleviating post-surgery pain [4]. Moreover, the introduction of molecularly predictive biomarkers has become a promising therapeutic option for patients with lung cancer [5, 6]. Nevertheless, to date, the exploration for early diagnostic and prognostic biomarkers for lung cancer remains a major clinical issue, as current knowledge in this area is still insufficient. Hence, exploring novel therapeutic biomarkers for patients with lung cancer is of great significance.
Circular RNAs (circRNAs) and microRNAs (miRNAs) belong to endogenous noncoding RNAs, which have recently garnered a lot of attention from researchers due to their involvement in a variety of biological processes [7–9]. CircRNAs are located in eukaryotic cells, and they are able to sponge miRNAs, working as transcriptionally modulatory molecules in various cellular processes, including the development of cancer [10, 11]. For example, circ-BPTF stimulated bladder cancer progression by means of the miR-31-5p/RAB27A axis [12]. Circ-RanGAP1 facilitated the metastasis and enhanced the invasive ability of gastric cancer by sponging miR-877-3p and modulating VEGFA levels [13]. Nonetheless, further efforts are necessary to investigate how circRNAs and miRNAs function in lung cancer development. Only a limited number of studies have reported on how circ_001621 takes part in cancer development. A study revealed that circ_001621 can enhance the proliferative and migratory abilities of osteosarcoma cells [14]. In this study, further investigation was conducted to elucidate the action of circ_001621 in lung cancer.
Gremlin 1 (GREM1) is from the bone morphogenic protein (BMP) antagonist family [15]. This family is characterized by its ability to modulate tissue differentiation, organogenesis, and body patterning [15]. Recent available data have revealed that GREM1 is upregulated in and clinically significant for human cancers, including colon cancer [16] and breast cancer [17, 18]. It is worth noting that GREM1 has been found to be upregulated in lung adenocarcinoma [19]. Still, further investigation is necessary to reveal the function of GREM1 in lung cancer progression.
The present study focused on investigating how circ_001621 operates in lung cancer pathogenesis. The potential mechanism of the circ_001621/miR-199a-3p/GREM1 axis in lung cancer development was also explored.
Material and methods
Samples and ethical approval
Thirty-six sets of lung cancer tissues and their corresponding adjacent noncancerous tissues were acquired at Wuhan Hospital of Traditional Chinese Medicine from 2019 to 2020. The samples excised from lung cancer patients were immediately frozen using liquid nitrogen, keeping them at –80°C until needed. All patients provided their consent in writing before the sample collection. The research has gained authorization from the ethics committee of Wuhan Hospital of Traditional Chinese Medicine.
Cell culture and transfection
The human lung cancer cell lines (Calu-3, A549, H1975, and PC9) as well as the human bronchial epithelial cell line BEAS-2B were bought from Procell (China). H1975 and PC9 were maintained in the RPMI 1640 medium (Procell), Calu-3 cells were cultivated in MEM (Procell), A549 cells were cultivated in Ham’s F-12K medium (Procell), and BEAS-2B cells were grown in DMEM (Procell). All media contained 10% FBS (Procell). All cell lines were incubated under the following conditions: 5% CO2 and 37°C. GenePharma (China) provided the si-circ_001621, miR-199a-3p inhibitor, miR-199a-3p mimic, si-GREM1, si-NC, inhibitor NC, and mimic-NC. The transfection concentration was 50 nM. The transfections were accomplished with the aid of Lipofectamine 2000 (Thermo Fisher Scientific, USA).
RNA extraction, RT-qPCR, and RNase R treatment
TRIzol reagent (Invitrogen, USA) was employed to extract the RNA from cells and tissues. The cDNA was then constructed by means of a Revert Aid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, USA). RT-qPCR was performed using a SYBR One Step RT-qPCR kit (Beijing Hooseen Biotech Co., Ltd, China) in the Bio-Rad CFX96 Real-Time PCR System (Bio-Rad, USA). The relative levels of RNA expression were computed by applying 2−ΔΔCt. The mRNA and miRNA internal controls were GAPDH and U6, respectively. The primer sequences are listed in Table I.
Table I
Real-time PCR primer sequences
Subcellular fractionation
The Cytoplasmic & Nuclear RNA Purification Kit (Amyjet Scientific, China) was used in this assay. Firstly, the cells were incubated for an hour in a fractionation buffer kept at ice-cold temperature. Subsequently, the cells were centrifuged to separate them into two main components: the supernatant and the pellet. The supernatant contained cytoplasmic lysate, whereas the pellet contained nuclear lysate. After RNA isolation, RT-qPCR was conducted to detect the circ_001621 expression in nuclear and cytoplasm with U6 and GAPDH as the controls.
CCK-8
H1975 and A549 cells were plated at a concentration of 1 × 104 cells/ml into 96-well plates. Once the cell density reached 70–80%, the corresponding oligonucleotides were transfected into the cells. The transfected cells were supplied with 10 µl of CCK-8 solution (Beyotime, China) at the time points of 0, 24, 48, and 72 h. The absorbance values were quantified at 450 nm using a microplate reader (BioTek, USA).
Caspase-3 activity assay
The caspase-3 colorimetric assay kit (Abcam, UK) was utilized in assessing the caspase-3 activity of the treated cells. In brief, the transfected cells were lysed with trypsin and centrifuged for 5 min at 600×g, 4°C. The supernatant was then collected and transferred to a 96-well plate. Afterwards, the cells were subjected to 4-hour incubation at 37°C with a reaction buffer that had caspase-3 substrate (Ac-DEVD-pNA). The absorbance (405 nm) was gauged using a microplate reader (BioTek).
Invasion detection
Transwell chambers (8-µm pores, Corning Incorporated, USA) containing Matrigel matrix (BD Biosciences, USA) were utilized to evaluate the capability of the lung cancer cells to invade. In the upper chamber, 200 µl of transfected lung cancer cells, with a concentration of 1 × 105 cells/ml, were loaded along with serum-free culture medium. The lower chamber was supplemented with 500 µl of complete culture medium, which served as an inducer for the upper cancer cells. Following a 24-hour incubation at 37°C, the lung cancer cells that invaded from the upper chamber to the lower chamber were fixed and then subjected to staining with 0.5% crystal violet (Beyotime, China). A microscope (Olympus, Japan) was utilized to photograph the visual fields.
Xenograft nude mouse model
The mice used for the in vivo study were 5-week-old female BALB/c nude mice. The tumor formation experiment was performed by injecting the left flank of each nude mouse with A549 cells (4 × 105) containing either the sh-circ_001621 lentiviral vector or its negative control (sh-NC) lentiviral vector (GenePharma, China). A caliper was utilized to gauge the tumor sizes weekly. Five weeks after injection, the mice were euthanized then their tumors were weighed. The Wuhan Hospital of Traditional Chinese Medicine – Institutional Animal Care and Use Committee authorized this experiment.
Dual-luciferase reporter experiment
The whole circ_001621 linear sequence was integrated into the pmirGLO vector to create the wild-type reporter vector, which included the predicted miR-199a-3p binding sites. The mutant was established by modifying the miR-199a-3p binding seed sequences from “ACUACUG” to “GTCGTCA”. The unchanged miR-199a-3p binding sequences on the GREM1-3’UTR were integrated into the pmirGLO vector to create the wild-type reporter vector. The GREM1-3’UTR mutant was constructed by modifying the miR-199a-3p binding sequences from “ACUACUG” to “GTCGTCA”. All luciferase reporters were constructed by Hongxun Biotech (Suzhou, China). In 96-well plates, H1975 and A549 were transfected with a combination of a wild-type/mutant circ_001621-3’UTR dual-luciferase reporter (100 ng) or GREM1-3’UTR reporter (100 ng), plus either a miR-199a-3p mimic or NC duplexes (50 nM). The assay was performed with six independent repetitions. After 48 h of transfection, the Firefly and Renilla luciferase activities were assessed by means of a Dual-GLO Luciferase Assay System Kit (Promega, USA) in a Fluorescence/Multi-Detection Microplate Reader (BioTek, USA).
RNA immunoprecipitation assay
This experiment was carried out by means of an EZ-Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, USA). H1975 and A549 cells successfully transfected with miR-199a-3p mimic were exposed to the RIP lysis buffer. Thereafter, the cell lysate was incubated with magnetic beads pre-coated with the anti-Ago2 or anti-IgG. The isolated RNA from the precipitation was subjected to reverse transcription to generate the cDNA. Lastly, RT-qPCR was done to assess circ_001621 and miR-199a-3p expression levels.
Western blotting
Total protein was acquired from A549 and H1975 treated with RIPA Lysis Buffer (Vazyme, China). Thereafter, 10% SDS-PAGE was utilized to isolate equal amounts of proteins from each group. The isolated proteins were subsequently placed onto PVDF membranes (Millipore, USA). The membranes were subjected to blocking with 5% skim milk for 2 h. Afterward, they were subjected to overnight incubation at 4°C with primary antibodies (anti-GREM1, 1 : 1000, Cat#: 4383, CST, USA or anti-GAPDH, 1 : 1000, Cat#: 5174, CST). On the following day, the primary antibodies were rinsed off with PBS three times, each time for 10 min. Afterward, the membranes were subjected to an hour of incubation with the second antibody (Abcam, UK). Lastly, the membranes underwent treatment with the Enhanced ECL luminescence detection kit (Vazyme) to visualize the proteins, which were subsequently examined using the FluorChem System.
Data analysis
The t-test and one-way ANOVA were applied accordingly to determine whether there was significant variance between or among groups, respectively. Pearson’s χ2 test was adopted to evaluate the relation between GREM1 and miR-199a-3p expression. The data are reported as mean ± standard deviation. A p-value < 0.05 was considered indicative of statistical significance. Each experiment was conducted with a minimum of three independent replicates.
Results
Circ_001621 was upregulated in lung cancer
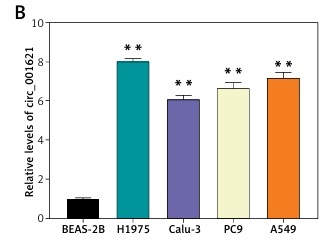
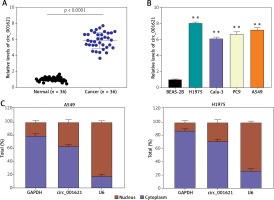
RT-qPCR was performed to validate the differential circ_001621 expression between tumoral and adjacent normal tissues. The lung tumor tissues exhibited a 6-fold rise in circ_001621 levels compared to normal tissues (Figure 1 A). Similar to the tissue level, we also quantified circ_001621 expression at the cellular level. The circ_001621 expression was significantly elevated in the lung cancer cell lines, particularly in H1975 and A549, where its expression was found to be approximately twice as high as in BEAS-2B (Figure 1 B). To further study the localization of circ_001621 within lung cancer cells, nuclei were isolated from A549 and H1975. RT-qPCR was conducted to assess circ_001621 distribution within the nucleus and cytoplasm. The results revealed that circ_001621 was predominately present within the cytoplasm of A549 and H1975 (Figure 1 C). Overall, our findings indicate that circ_001621 is considerably upregulated in lung cancer.
Figure 1
Circ_001621 was upregulated in lung cancer. A – Circ_001621 is highly expressed in lung cancer tissues by RT-qPCR. B – Circ_001621 mRNA expression in human lung cancer cell lines A549, Calu-3, PC9 and H1975, and human bronchial epithelial cell line BEAS-2B by qRT-PCR. **P < 0.001 versus BEAS-2B cells. C – Localization of circ_001621 in cytoplasm and nucleus of A549 and H1975 cells. Data are presented as mean ± SD
Circ_001621 downregulation inhibited lung cancer cell malignancy
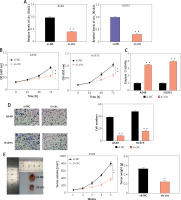
In order to explore how circ_001621 affected lung cancer development, a specific siRNA of circ_001621 (si-circ) was synthesized and transfected into A549 and H1975. Si-circ resulted in effective knockdown in A549 and H1975, with a knockdown efficiency of over 75% (Figure 2 A). The influence of circ_001621 on cell viability was ascertained through a CCK-8 experiment. It was evident that the si-circ transfection resulted in a 20% decline in OD (450 nm) at 96 h (Figure 2 B), indicating that circ_001621 had a positive effect on cell viability. Subsequently, caspase-3 activity was assessed to validate whether circ_001621 affected lung cancer cell apoptosis. Upon transfecting the cells with si-circ, the caspase-3 activity in A549 and H1975 increased by 3.5-fold and 4-fold, respectively (Figure 2 C). This suggests that circ_001621 has an antagonistic effect on lung cancer cell apoptosis. The invasion ability of H1975 and A549 was evaluated by means of a transwell experiment. The results demonstrated that si-circ transfection considerably reduced their invasion ability (Figure 2 D), indicating that circ_001621 was able to promote lung cancer cell invasion. A nude mouse tumorigenicity experiment was conducted to directly observe how circ_001621 influenced tumorigenicity. It was observed that the sh-circ treatment considerably diminished tumor size and tumor weight (Figure 2 E), suggesting the strong tumorigenicity of circ_001621 in lung cancer. Overall, circ_001621 appears to facilitate lung cancer development.
Figure 2
Circ_001621 contributed to lung cancer development. A – Transfection efficiency of si-circ_001621 in A549 and H1975 cells by RT-qPCR. B – Cell viability was detected by CCK-8 assay. C – Cell apoptosis was detected by caspase-3 activity assay. D – Cell invasion was detected by transwell assay. E – Xenograft nude mouse model was designed to detect the effect of circ_001621 on lung cancer growth in vivo. *P < 0.05, **p < 0.001 versus si-NC or sh-NC. Data are presented as mean ± SD
Circ_001621 could sponge miR-199a-3p in lung cancer progression
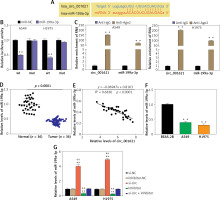
StarBase was employed to predict the potential target miRNA of circ_001621. The outcomes revealed that circ_001621 has potential to target miR-199a-3p, which matched circ_001621 in its seed sequence (Figure 3 A). The binding of circ_001621 with miR-199a-3p was verified by means of the dual-luciferase reporter experiment. Relative to other groups, the fluorescence value in the WT+miR-199a-3p showed a ~60% decline (Figure 3 B), indicating a stable binding effect. The target relationship of circ_001621 with miR-199a-3p was further validated through an RIP experiment. It was found that circ_001621 was upregulated, and there was enrichment of miR-199a-3p in the RIP product (Figure 3 C). Based on the target relationship, the expression of miR-199a-3p in tumors was quantified. In contrast to circ_001621, miR-199a-3p exhibited a decline of over 50% in tumors relative to normal tissues (Figure 3 D). The expression correlation revealed that as the miR-199a-3p RNA levels declined, circ_001621 levels increased (Figure 3 E). Similarly, miR-199a-3p exhibited downregulation in A549 and H1975, in contrast to what was observed for circ_001621 (Figure 3 F). The si-circ treatment demonstrated its ability to upregulate the miR-199a-3p RNA level, and it even alleviated the inhibitory effect caused by miR-199a-3p inhibitor (Figure 3 G), suggesting that circ_001621 has an endogenic target relationship with miR-199a-3p. These findings suggest that circ_001621 can target miR-199a-3p.
Figure 3
Circ_001621 could sponge miR-199a-3p in lung cancer progression. A – Predicted binding sites between miR-199a-3p and circ_001621. B – The target relationship between miR-199a-3p and circ_001621 was verified by dual-luciferase reporter assay. **P < 0.001 versus mimic-NC. C – Relative enrichment of circ_0032463 in anti-IgG and anti-Ago2 was detected by RIP assay. **P < 0.001 versus anti-IgG group. D – MiR-199a-3p expression in lung cancer tissues and non-tumor tissues by RT-qPCR. E – Expression of miR-199a-3p had a negative correlation with circ_001621 expression in A549 and H1975 cells. F – MiR-199a-3p expression in human lung cancer cell lines A549 and H1975, and human bronchial epithelial cell line BEAS-2B by RT-qPCR. **P < 0.001 versus BEAS-2B cells. G – Transfection efficiency of miR-199a-3p mimic by RT-qPCR in A549 and H1975 cells. **P < 0.001 versus si-NC. ##P < 0.001 versus inhibitor-NC. &&P < 0.001 versus si-circ + inhibitor. Data are presented as mean ± SD
Circ_001621 exerted its oncogenic influence on lung cancer cells by inhibiting miR-199a-3p
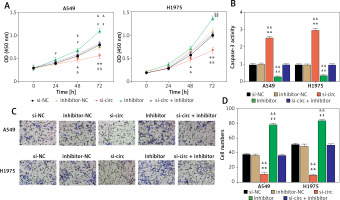
With regard to the circ_001621—miR-199a-3p target relationship, miR-199a-3p was considered to be an essential downstream factor in the regulation of lung cancer development by circ_001621. As depicted in Figure 4 A, the poor expression of miR-199a-3p enhanced the A549 and H1975. However, this promotion could be reversed by si-circ treatment. Caspase-3 activity was measured to assess the role of miR-199a-3p in antagonizing apoptosis in lung cancer cells. It was observed that, under si-circ treatment, caspase-3 activity increased 3 times and 2.5 times in A549 and H1975, respectively. The knockdown of miR-199a-3p repressed caspase-3 activity, which was then restored by si-circ (Figure 4 B). Similarly, cell invasion ability was assessed. The inhibitor treatment strengthened the invasion ability of A549 and H1975, while si-circ counteracted this promotion (Figure 4 C). Therefore, circ_001621 exerts its oncogenic effect on lung cancer cells by inhibiting miR-199a-3p.
Figure 4
The oncogenic effect of circ_001621 on lung cancer cells was exerted via inhibiting miR-199a-3p. A – Cell viability was detected by CCK-8 assay. B – Cell apoptosis was detected by caspase-3 activity assay. C, D – Cell invasion was detected by transwell assay. *P < 0.05, **p < 0.001 versus si-NC. #P < 0.05, ##p < 0.001 versus inhibitor-NC. &P < 0.05, &&p < 0.001 versus si-circ + inhibitor. Data are presented as mean ± SD
MiR-199a-3p targeted GREM1 in lung cancer cells
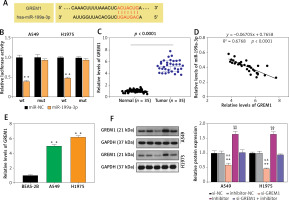
TargetScan was employed to predict the downstream target mRNA of miR-199a-3p. According to the prediction, miR-199a-3p could target and bind to position 2187-2194 of the GREM1 3’UTR (Figure 5 A). The dual-luciferase reporter experiment demonstrated that co-transfecting miR-199a-3p mimic with wild-type plasmids resulted in approximately a 60% reduction in fluorescence value, indicating the binding between them (Figure 5 B). RT-qPCR was conducted to quantify GREM1 RNA levels in tumor tissues. It revealed that GREM1 RNA expression was upregulated approximately 5-fold in lung cancer tumor tissues (Figure 5 C). The RNA correlation analysis indicated that the expression of GREM1 mRNA increased as the miR-199a-3p RNA level declined (Figure 5 D). Compared to BEAS-2B, the GREM1 mRNA levels in A549 and H1975 were increased by more than twofold (Figure 5 E). The outcomes of western blotting showed that the presence of si-GREM1 resulted in downregulation of the GREM1 protein level, while the miR-199a-3p inhibitor could raise GREM1 protein levels back to the normal range (Figure 5 F). This indicates that the miR-199a-3p inhibitor could increase the endogenic GREM1 expression. These results substantiate that miR-199a-3p targets GREM1.
Figure 5
MiR-199a-3p targeted GREM1 in lung cancer cells. A – Predicted binding sites between miR-199a-3p and GREM1. B – The target relationship between miR-199a-3p and GREM1 was verified by dual-luciferase reporter assay. **P < 0.001 versus mimic-NC. C – GREM1 mRNA expression in lung cancer tissues and non-tumor tissues by RT-qPCR. D – Expression of miR-199a-3p had a negative correlation with GREM1 expression in A549 and H1975 cells. E – GREM1 mRNA expression in human lung cancer cell lines A549 and H1975, and human bronchial epithelial cell line BEAS-2B by RT-qPCR. **P < 0.001 versus BEAS-2B cells. F – Relative protein expression of GREM1 in A549 and H1975 was detected by western blotting assay. **P < 0.001 versus si-NC. ##P < 0.001 versus inhibitor-NC. &&P < 0.001 versus si-GREM1 + inhibitor. Data are presented as mean ± SD
MiR-199a-3p inhibited lung cancer cell activity through suppression of GREM1
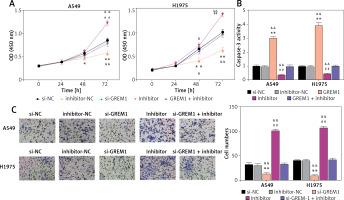
As shown in Figure 6 A, si-GREM1 transfection significantly diminished the OD (450 nm) value in A549 and H1975. Moreover, the upregulation of its endogenic expression, which was induced by the miR-199a-3p inhibitor, restored that inhibitory impact. After transfecting si-GREM1, the caspase-3 activity increased 3 times and 4 times in A549 and H1975, respectively. In contrast to si-GRME1, the miR-199a-3p inhibitor treatment reduced the caspase-3 activity by about 50%. Furthermore, this reduction could be restored to normal levels under si-GREM1 treatment, indicating that GREM1 was capable of counteracting apoptosis in lung cancer tumor cells (Figure 6 B). With regard to tumor cell invasion ability, si-GREM1 dramatically reduced invasion ability, while miR-199a-3p inhibitor restored it to normal levels in both A549 and H1975 (Figure 6 C). Overall, these findings suggest that GREM1 facilitates lung cancer development.
Figure 6
MiR-199a-3p inhibited lung cancer cell activity by suppressing GREM1. A – Cell viability was detected by CCK-8 assay. B – Cell apoptosis was detected by caspase-3 activity. C – Cell invasion was detected by transwell assay. *P < 0.05, **p < 0.001 versus si-NC. ##P < 0.001 versus inhibitor-NC. &P < 0.05, &&p < 0.001 versus si-GREM1 + inhibitor. Data are presented as mean ± SD
Discussion
A growing body of evidence has revealed that circRNAs actively partake in a variety of biological processes, particularly in tumorigenesis [20–22]. Nevertheless, much remains to be uncovered about circ_001621’s underlying regulatory mechanism in lung cancer. Herein, we focused on the role of circ_001621 in lung cancer cell viability, apoptosis, invasion, and cancer growth, both in vitro and in vivo. Consequently, we observed the dramatic upregulation of circ_001621 expression in both lung cancer cells and tissues. In addition, circ_001621 exhibited more stability than the linear RNA as well as resistance against RNase R degradation. The in vitro biological function experiments demonstrated that circ_001621 silencing could reduce the invasive ability and viability of lung cancer cells while promoting their apoptosis. Furthermore, the anti-tumor activity of the silenced circ_001621 was further substantiated by the results of the in vivo nude mouse experiment. It was also verified that the miR-199a-3p/GREM1 axis could interact with circ_001621 in lung cancer development. This axis holds promise as a prospective biomarker in the future for lung cancer treatment.
We are the first to detect abundant circ_001621 expression in lung cancer cells and tissues. As its enrichment was detected in lung cancer progression, circ_001621 demonstrated its role as an oncogenic circRNA. Hence, its downregulation, which was affected by si-circRNA, resulted in repressed cell viability and invasion as well as enhanced apoptosis. Our in vivo assay further suggested that the downregulation of circ_001621 was capable of inhibiting lung cancer growth. Supporting our findings, a recent study [14] reported that circ_001621 was among the significantly upregulated circRNAs identified through circRNA microarrays in osteosarcoma tissues. Furthermore, its elevated expression is correlated with the advanced stages of osteosarcoma. More importantly, this study demonstrated that elevated circ_001621 expression is associated with shorter overall survival [14]. Despite the limited available evidence regarding the characterization of circ_001621 characterization, this research, in conjunction with previous findings of Ji et al. [14], indicates that circ_001621 may function as an active pro-tumor factor in cancer progression.
The most recognized property of circRNAs is their ability to be a “miRNA sponge.” They could bind and restrain the function of miRNAs, hence preventing their target genes from being degraded [23–26]. Herein, we discovered that miR-199a-3p could be sponged by circ_001621. The statistical analysis also revealed that the expression levels of circ_001621 and miR-199a-3p in lung cancer tissues were negatively correlated. We then tried to elucidate how miR-199a-3p operates in lung cancer cells. The results showed that miR-199a-3p could impede lung cancer progression by reducing lung cancer cell viability and invasive ability while also stimulating apoptosis. In previous data, miR-199a-3p was also revealed to act as an anti-tumor factor in a variety of cancers. For instance, Callegari et al. [27] observed that miR-199a-3p regulated the MTOR and PAK4 pathways and hampered the growth of tumor in a hepatocellular carcinoma transgenic mouse model. A similar observation has been reported in ovarian cancer [28]. Our findings were in line with earlier research. To delve deeper into the interaction of miR-199a-3p with circ_001621 in lung cancer cells, we executed a series of rescue experiments. The outcomes indicated that the suppressive influence of the silenced circ_001621 on lung cancer cell development could be counteracted by the introduction of a miR-199a-3p inhibitor.
Last but not least, we identified that circ_001621 sponged miR-199a-3p to upregulate GREM1. GREM1 encodes a member of the BMP antagonist family, which partakes in the modulation of organogenesis, body patterning, and tissue differentiation [15]. In this regard, it is evident that GREM1 may have an essential role in regulating various cellular activities, which could be related to the initiation and growth of cancer. Numerous previous studies about GREM1 have reported its association with the progression of different cancers. In colon cancer, GREM1 downregulation dramatically restrained colon cancer cell proliferation and migration through the suppression of the phosphorylation levels of the BMP downstream signal Smad1, vascular endothelial growth factor downstream signal matrix metallopeptidase 2, and metastasis-related factor C-X-C motif chemokine ligand 12 expression [16]. As for breast cancer, GREM1 contributed to metastasis by triggering the STAT3-MMP13 signaling pathway activation [17]. The elevated GREM1 expression has been implicated in lung adenocarcinoma, and it was also reported to stimulate the proliferation and growth of normal lung cells [19]. In line with these findings, our data also indicated overexpression of GREM1 in lung cancer tissues. Moreover, we observed that GREM1 enhanced the invasive ability and viability of lung cancer cells but repressed apoptosis. MiR-199a-3p was found to inhibit these effects, while circ_001621 was observed to promote them.
This study has limitations. The migratory ability of the lung cancer cells was not explored although the invasion was verified by transwell assay. Furthermore, a comprehensive exploration of the BMP signaling pathway is warranted to improve our understanding of the regulatory mechanism underlying circ_001621/miR-199a-3p/GREM1 in lung cancer development.
In conclusion, this study confirms significant enrichment of circ_001621 in lung cancer, and circ_001621 knockdown impaired the malignancy of lung cancer cells in vivo and in vitro. Moreover, this study demonstrated that circ_001621 can bind with miR-199a-3p and boost the levels of its target GREM1. Hence, our study has uncovered a novel regulatory axis consisting of circ_001621/miR-199a-3p/GREM1 in lung cancer development, offering novel insights and prospective targets for lung cancer diagnosis and treatment.