Pancreatic adenocarcinoma (PaCa) is a malignant neoplasm that arises from the exocrine component of the pancreas, which is responsible for producing digestive enzymes [1]. Risk factors of PaCa may include chronic pancreatitis, diabetes, smoking, and obesity, while it is more common in older individuals and tends to have a slight male predominance [2].
Several circular RNAs (circRNAs) such as linc00152, circRNA_0030235, circRNA_0001649, circRNA_001569, and circ_0005397 have been reported to play a pivotal role in PaCa by the modulation of downstream pathways related to metabolism, inflammation, cell proliferation and cell apoptosis [3, 4]. miR-326 may target and downregulate specific signaling pathways known to be involved in PaCa progression, such as Hedgehog signaling [5]. Similarly, miR-216b also acts as a tumor suppressor [6]. It may inhibit cell growth, induce apoptosis, and potentially prevent metastasis in PaCa patients. miR-216b may target and negatively regulate PaCa-related oncogenes or modulate signaling pathways involved in the pathogenesis of PaCa progression, contributing to its tumor-suppressive effects [7, 8]. Therefore, the expression levels of miR-216b could serve as a prognostic marker.
Pyruvate dehydrogenase kinase 2 (PDK2) serves as a regulator of the pyruvate dehydrogenase complex (PDC), thus being pivotal in cellular metabolic processes which influence the pathogenesis of various cancers [9]. However, although studies to date have not reported a direct association between PDK2 and PaCa, the target molecule of PDK2, i.e. Akt, has been reported to participate in the pathogenesis of PaCa [10]. Considering the pivotal role of PDK2 in glycolysis [11], we reasoned that investigation of the regulation of PDK2 will contribute to the elucidation of pathogenesis of the disease. Furthermore, non-coding RNAs such as lincRNAs and circRNAs have been reported to be involved in the oncogenesis of pancreatic ductal adenocarcinoma [4], and PDK2 was also reported to be regulated by hsa_circ_0005397 [12]. Therefore, this study focused on the non-coding RNAs as upstream regulators of PDK2. In this study, we proposed that the non-coding RNAs linc00152 and hsa_circ_0005397 may interact with their target miRNAs, specifically miR-326 and miR-216b, to modulate the expression of PDK2, a crucial enzyme in cellular metabolism. The circ_0005397/miR-326/PDK2 and linc00152/miR-216b/PDK2 signaling axes represent a sophisticated layer of post-transcriptional regulation, contributing to the metabolic reprogramming that typifies cancer cells.
In this study, we recruited 103 patients with pancreatic cancer as the PaCa group (N = 103) and 103 healthy individuals as the control group (N = 103). Peripheral blood samples were collected from both groups for subsequent analysis. Moreover, PaCa tissues and PaCa adjacent tissues were collected from each PaCa patient for subsequent analysis. All protocols were approved by the Ethics Committees of Liaoning Cancer Hospital & Institute (IRB: LN-ZL-212308) and all experiments were performed in strict accordance with the latest version of the Declaration of Helsinki. All participants signed informed consent before initiation of this study.
PaCa cells were used to establish cell models in this study. In cell model 1, 4 groups were established: an NC group, a linc00152 group, a circ_000539 group, and a linc00152 + circ_000539 group. In cell model 2, 4 groups were also established: a scramble siRNA group, a linc00152 siRNA group, a circ_000539 siRNA group, and a linc00152 siRNA + circ_000539 siRNA group.
The total RNA content was extracted from the samples (blood, tissue and cells) using the TRIzol reagent kit (Invitrogen). The purity and concentration of total RNA were determined by optical density measurements. The relative expression levels of target genes were determined using the 2–ΔΔCt method. GAPDH and U6 were utilized as the internal reference gene for real-time PCR.
Target prediction analysis was performed to identify the molecular interactions between linc00152, linc00152, miR-326, miR-216b, and PDK2 mRNA. To further validate the molecular interactions between miR-326 and linc00152, miR-216b and circ_0005397, miR-326 and PDK2 mRNA, and miR-216b and PDK2 mRNA, luciferase assays were performed.
Statistical analysis was performed using GraphPad Prism 7 (GraphPad Software, Inc.). The results were expressed as mean ± standard deviation (SD). The differences of circRNA expression and lncRNA expression between PaCa tissues and PaCa adjacent tissues, or between the PaCa group and normal control group, were compared using t-tests. The comparisons between the luciferase assay cell groups or multiple cell model groups were performed using one-way ANOVA, corrected by Tukey’s post-hoc test. A p-value < 0.05 was considered statistically significant.
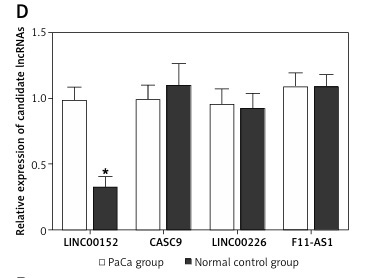
As shown in Table I, no significant differences were observed with respect to demographic parameters such as age and gender distribution, family history, smoking habit and diabetes, thus excluding these parameters as potential confounding factors. By matching the results from Figures 1 A and B which respectively indicated dysregulated circRNAs, we found that circ_0005397 was up-regulated in both PaCa tissues compared with adjacent non-cancerous tissues, and in PaCa patients compared with healthy control individuals, which suggested its diagnostic potential as a non-invasive biomarker. Figures 1 C and D compare expression of candidate lncRNAs in PaCa versus adjacent non-cancerous tissues. The results suggest that linc00153 may play a pivotal role in the pathogenesis of PaCa, since it is higher in PaCa tissues compared with PaCa adjacent tissues, as well as in PaCa patients compared with normal participants.
Table I
Demographic information of all participants
Figure 1
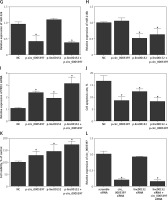
A – Comparison of candidate circRNA relative expression levels in PaCa tissues and in PaCa adjacent tissues. B – Comparison of candidate circRNA relative expression levels in plasma samples collected from PaCa patients and healthy control individuals. C – comparison of candidate lncRNA relative expression levels in PaCa tissues and in PaCa adjacent tissues. D – Comparison of candidate lncRNA relative expression levels in plasma samples collected from PaCa patients and healthy control individuals. E – Relative expression of circ_005397 in NC, p-linc00152, p-circ_0005397 and p-linc00152 + p-circ_0005397 group. F – Relative expression of linc00152 in NC, p-linc00152, p-circ_0005397 and p-linc00152 + p-circ_0005397 group. G – Relative expression of miR-326 in NC, p-linc00152, p-circ_0005397 and p-linc00152 + p-circ_0005397 group. H – Relative expression of miR-216b in NC, p-linc00152, p-circ_0005397 and p-linc00152 + p-circ_0005397 group. I – Relative expression of PDK2 mRNA in NC, p-linc00152, p-circ_0005397 and p-linc00152 + p-circ_0005397 group. J – Apoptosis rate of the NC, p-linc00152, p-circ_0005397 and p-linc00152 + p-circ_0005397 group. K – MTT assay results of cell proliferation in NC, p-linc00152, p-circ_0005397 and p-linc00152 + p-circ_0005397 group. L – Relative expression of circ_005397 in scramble siRNA, lin00152 siRNA, circ_0005397 siRNA and linc00152 siRNA + circ_0005397 siRNA group. M – Relative expression of linc00152 in scramble siRNA, lin00152 siRNA, circ_0005397 siRNA and linc00152 siRNA + circ_0005397 siRNA group. N – Relative expression of miR-326 in scramble siRNA, lin00152 siRNA, circ_0005397 siRNA and linc00152 siRNA + circ_0005397 siRNA group. O – Relative expression of miR-216b in scramble siRNA, lin00152 siRNA, circ_0005397 siRNA and linc00152 siRNA + circ_0005397 siRNA group. P – Relative expression of PDK2 mRNA in scramble siRNA, lin00152 siRNA, circ_0005397 siRNA and linc00152 siRNA + circ_0005397 siRNA group. Q – Apoptosis rate of the scramble siRNA, lin00152 siRNA, circ_0005397 siRNA and linc00152 siRNA + circ_0005397 siRNA group. R – MTT assay results of cell proliferation in scramble siRNA, lin00152 siRNA, circ_0005397 siRNA and linc00152 siRNA + circ_0005397 siRNA group

Sequence analysis identified specific binding sites between circ_005397 and miR-326 (Supplementary Figure S1 A), linc00152 and miR-216b (Supplementary Figure S1 B), PDK2 mRNA and miR-326 (Supplementary Figure S1 C), and PDK2 mRNA and 216b (Supplementary Figure S1 D) by sequence analysis. The corresponding luciferase assays performed in PaCa and PANC-1 cells all demonstrated evidently suppressed luciferase activities in cells co-transfected with wild-type circ_005397 and miR-326 mimics (Supplementary Figure S1 A), or wild-type linc00152 and miR-216b mimics (Supplementary Figure S1 B), or wild-type PDK2 mRNA 3’UTR and miR-326 (Supplementary Figure S1 C), or wild-type PDK2 mRNA 3’UTR and miR-216b (Supplementary Figure S1 D). Therefore, the above experiments suggested the establishment of two potential signaling pathways: the circ_0005397/miR-326/PDK2 mRNA pathway and the linc0152/miR-216b/PDK2 mRNA pathway.
We also studied the effects of the dysregulation of circ_005397/linc00152 on the modulation of the above-established signaling pathways. The highly expressed circ_005397 by p-circ_005397 (Figure 1 E), as well as the highly expressed linc00152 by p-linc00152 (Figure 1 F) validated the successful transfection of these plasmids. Moreover, both the p-circ_005397 and the co-transfection (p-circ_005397 + p-linc00152) group exhibited reduced miR-326 levels (Figure 1 G), and the p-linc00152 and the co-transfection group exhibited reduced miR-216b levels (Figure 1 H). However, PDK2 mRNA was significantly increased in the p-circ_005397 group, p-linc00152 group and the co-transfection group, suggesting its role as a shared target of the circ_005397/miR-326 axis and the linc00152/miR-216b axis. Meanwhile, the apoptosis rate was the highest in the NC group (Figure 1 I) while the proliferation rate was the lowest in the NC group (Figure 1 J), suggesting that overexpression of circ_005397 or linc00152 could suppress cell apoptosis and promote cell proliferation. It is also noteworthy that the co-transfection of p-circ_005397 and p-linc00152 showed a stronger effect in modulating these cell processes.
Moreover, the successful knockdown of circ_005397 (Figure 1 L) and linc00152 (Figure 1 M) both led to significant down-regulation of PDK2 mRNA (Figure 1 P), via the up-regulation of miR-326 (Figure 1 N) and miR-216b (Figure 1 O), respectively. Also, the knockdown of circ_005397 and linc00152 significantly promoted cell apoptosis (Figure 1 Q) and inhibited the cell proliferation rate (Figure 1 R). Therefore, all the above observations confirmed that the circ_0005397/miR-326/PDK2 mRNA pathway and the linc00152/miR-216b/PDK2 mRNA pathway could promote the oncogenesis of PaCa.
circ_0005397 was found to be implicated in the modulation of various tumor-related pathways through its interaction with target miRNAs such as miR-26b, miR-125a, miR-330, and miR-382. In a previous study which investigated the role of circ_0005397 in PaCa [13], the level of circ_0005397 was found to be elevated in PaCa tissues and corresponding cell lines, and the targeted suppression of circ_0005397 can hinder the proliferation, invasion, and migration of pancreatic cells, underscoring its pivotal role in PaCa via the sponging of oncogenic miRNAs. Therefore, circ_0005397 is recognized as a potential therapeutic target in PaCa treatment. In this study, we verified that circ_0005397 could participate in the pathogenesis of PaCa oncogenesis by regulating miR-26b and its downstream target gene, PDK2. Similar to previous research, we also found that the level of circ_0005397 is up-regulated in PaCa patients compared with healthy individuals.
linc00152 was initially reported to be overexpressed in gastric cancer [14], exhibiting its role as an oncogene in PaCa [15]. Specifically, linc00152 was found to be associated with cell cycle arrest, enhanced cell migration and invasion, and cell apoptotic processes in gastric cancer [16]. Moreover, a previous study by Yuan et al. also observed the overexpression of linc00152 in human PaCa tissue compared with healthy individuals [4], which is similar to the findings of our study. Yuan et al. also validated their observations in PaCa cell lines, confirming the association between linc00152 upregulation and increased cell proliferation, invasion, and migration by sponging miR-150 [4]. In our study, a different miRNA, miR-216b, was identified to be sponged by linc00152 by performing sequence analysis and luciferase assays. A shared target gene, PDK2 mRNA, was also identified to be regulated by the circRNA_0005397/miR-326 axis and linc00152/miR-216b axis in the pathogenesis of PaCa oncogenesis.
Although no direct correlation of PDK2 and PaCa has been reported, Akt, a molecule phosphorylated and activated by PDK2, has been proved to be overexpressed in subjects with PaCa in many studies [10, 17]. In this study, PDK2 mRNA was found to be targeted by both the circRNA_0005397/miR-326 axis and the linc00152/miR-216b axis, exhibiting significantly increased expression levels in PaCa tissues. Also, the PaCa cell proliferation increased correspondingly with the promoted risk of oncogenesis. The result of this present study is in line with the previous study about the relationship between PDK2 and circRNA_0005397, and this further confirmed the role of PDK2 in the pathogenesis of oncogenesis in PaCa. It suggests that PDK2 and its regulators could potentially serve as biomarkers for the early diagnosis, prognosis, and prediction of response to chemotherapy in PaCa.
However, this study is limited by the relatively small sample size. Also, we need to further validate our results in an independent cohort, thus strengthening the evidence for the role of circ_0005397/miR-326 and linc00152/miR-216b in pancreatic adenocarcinoma.
In conclusion, in this study, we found that circ_0005397 and linc00152 could interact with miR-326 and miR-216b, respectively, to modulate the expression of PDK2 in PaCa. The circ_0005397/miR-326/PDK2 and linc00152/miR-216b/PDK2 signaling axes can therefore promote the oncogenesis of PaCa.