Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Section Editors
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
CLINICAL RESEARCH
Causal associations of circulating inflammatory proteins with sepsis: a two-sample Mendelian randomization study
1
Department of Critical Care Medicine, Wuxi No. 2 People’s Hospital, Jiangnan University Medical Center, Wuxi, China
2
Department of Critical Care Medicine, Aheqi County People’s Hospital, Xinjiang, China
3
Department of Critical Care Medicine, Yuncheng Central Hospital, The Eighth Affiliated Medical College of Shanxi Medical University, Yuncheng, China
4
Department of Gynaecology and Obstetrics, Wuxi Maternity and Child Health Care Hospital, Affiliated Women’s Hospital of Jiangnan University, Wuxi, China
Submission date: 2023-12-15
Final revision date: 2024-02-04
Acceptance date: 2024-02-11
Online publication date: 2024-12-13
Corresponding author
Jia Wu
Department of Gynaecology and Obstetrics, Wuxi Maternity and Child Health Care Hospital Affiliated Women’s Hospital of Jiangnan University, Wuxi 214002, China No. 48 Huaishu Lane Liangxi District Wuxi, Jiangsu 214002, China
Department of Gynaecology and Obstetrics, Wuxi Maternity and Child Health Care Hospital Affiliated Women’s Hospital of Jiangnan University, Wuxi 214002, China No. 48 Huaishu Lane Liangxi District Wuxi, Jiangsu 214002, China
KEYWORDS
TOPICS
ABSTRACT
Introduction:
Sepsis arises from dysregulated inflammation in response to infection, precipitating organ dysfunction. Circulating inflammatory mediators likely contribute to sepsis pathogenesis, but their precise roles remain unclear. We aimed to evaluate potential causal impacts of inflammatory proteins on sepsis risk.
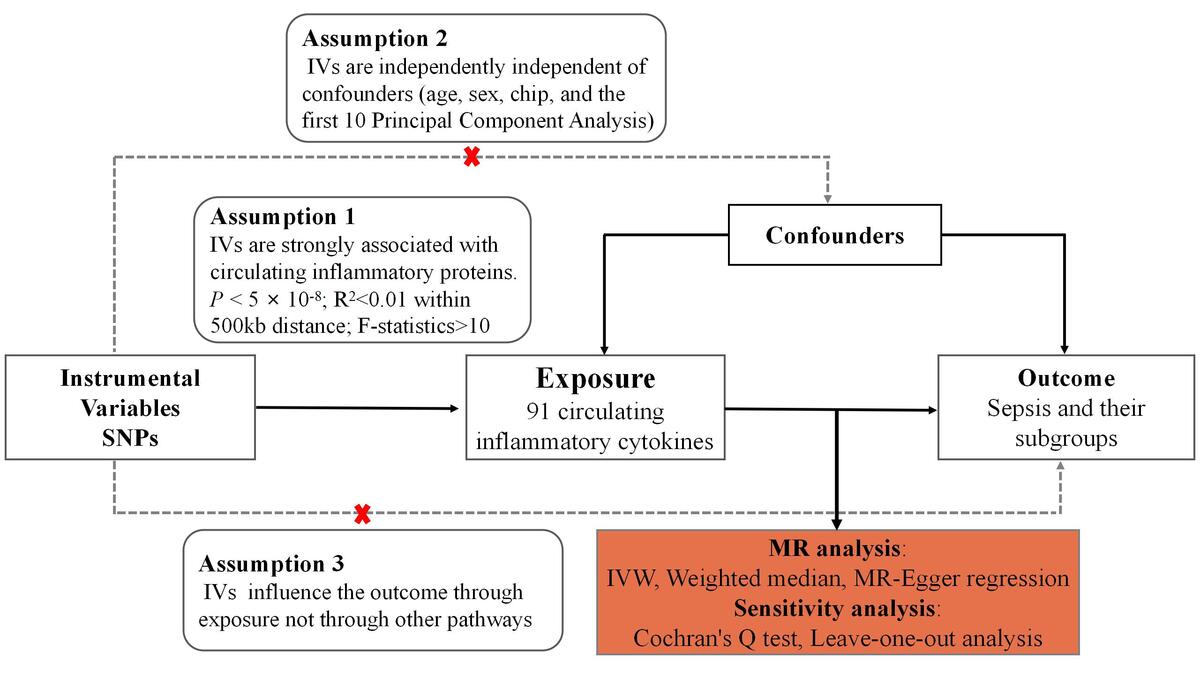
Material and methods:
We performed a two-sample Mendelian randomization study evaluating causal associations for 91 inflammatory proteins with sepsis risk. Genetic instruments were derived from a published genome-wide association study of plasma proteins. Sepsis outcome data were obtained from the UK Biobank and FinnGen cohort. Inverse-variance weighted analysis was conducted along with several sensitivity analyses.
Results:
The analyses identified significant causal associations for several inflammatory proteins with sepsis risk. TRAIL exhibited a causal effect on increased overall sepsis risk (OR = 1.10, 95% CI: 1.03–1.17). CCL28 showed a causal link to higher 28-day sepsis mortality in critical care (OR = 3.32, 95% CI: 1.18–9.29). CCL4 demonstrated a causal association with increased 28-day sepsis mortality (OR = 1.81, 95% CI: 1.06–1.31). Meanwhile, beta-NGF was found to be causally protective for sepsis (OR = 0.77, 95% CI: 0.60–0.99), and TNFB also showed a causally protective effect (OR = 0.95, 95% CI: 0.91–1.00).
Conclusions:
Our study elucidates roles of inflammatory mediators in sepsis pathogenesis. The identified proteins may serve as biomarkers or therapeutic targets. TRAIL signaling inhibition may hold promise for future clinical translation given its causal links to increased sepsis risk and mortality. CCL28 and CCL4 also represent potential immunological drivers of sepsis mortality worthy of further investigation. Meanwhile, the neurotrophins beta-NGF and TNFB emerged as having protective effects in sepsis that could be therapeutically augmented. Further experimental validation is warranted to confirm the observed causal relationships. Our findings provide targets for future mechanistic and clinical examination to impact patient prognosis.
Sepsis arises from dysregulated inflammation in response to infection, precipitating organ dysfunction. Circulating inflammatory mediators likely contribute to sepsis pathogenesis, but their precise roles remain unclear. We aimed to evaluate potential causal impacts of inflammatory proteins on sepsis risk.
Material and methods:
We performed a two-sample Mendelian randomization study evaluating causal associations for 91 inflammatory proteins with sepsis risk. Genetic instruments were derived from a published genome-wide association study of plasma proteins. Sepsis outcome data were obtained from the UK Biobank and FinnGen cohort. Inverse-variance weighted analysis was conducted along with several sensitivity analyses.
Results:
The analyses identified significant causal associations for several inflammatory proteins with sepsis risk. TRAIL exhibited a causal effect on increased overall sepsis risk (OR = 1.10, 95% CI: 1.03–1.17). CCL28 showed a causal link to higher 28-day sepsis mortality in critical care (OR = 3.32, 95% CI: 1.18–9.29). CCL4 demonstrated a causal association with increased 28-day sepsis mortality (OR = 1.81, 95% CI: 1.06–1.31). Meanwhile, beta-NGF was found to be causally protective for sepsis (OR = 0.77, 95% CI: 0.60–0.99), and TNFB also showed a causally protective effect (OR = 0.95, 95% CI: 0.91–1.00).
Conclusions:
Our study elucidates roles of inflammatory mediators in sepsis pathogenesis. The identified proteins may serve as biomarkers or therapeutic targets. TRAIL signaling inhibition may hold promise for future clinical translation given its causal links to increased sepsis risk and mortality. CCL28 and CCL4 also represent potential immunological drivers of sepsis mortality worthy of further investigation. Meanwhile, the neurotrophins beta-NGF and TNFB emerged as having protective effects in sepsis that could be therapeutically augmented. Further experimental validation is warranted to confirm the observed causal relationships. Our findings provide targets for future mechanistic and clinical examination to impact patient prognosis.
REFERENCES (29)
1.
Rudd KE, Johnson SC, Agesa KM, et al. Global, regional, and national sepsis incidence and mortality, 1990-2017: analysis for the Global Burden of Disease Study. Lancet (London, England) 2020; 395: 200-11.
2.
Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 2016; 315: 801-10.
3.
Siampa VN, Abadi S, Aman AM, et al. Association between severity of sepsis and thyroid function profile. Acta Biomed 2023; 94: e2023239.
4.
Delano MJ, Ward PA. Sepsis-induced immune dysfunction: can immune therapies reduce mortality? J Clin Investig 2016; 126: 23-31.
5.
Marshall JC. Why have clinical trials in sepsis failed? Trends Mol Med 2014; 20: 195-203.
6.
Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 2014; 23: R89-98.
7.
Relton CL, Davey Smith G. Mendelian randomization: applications and limitations in epigenetic studies. Epigenomics 2015; 7: 1239-43.
8.
Wu J, Zhang X, Wu D, et al. Evaluation of causal associations between interleukin-18 levels and immune-mediated inflammatory diseases: a Mendelian randomization study. BMC Med Genom 2023; 16: 306.
9.
Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018; 7: e34408.
10.
Burgess S, Scott RA, Timpson NJ, et al. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol 2015; 30: 543-52.
11.
Skrivankova VW, Richmond RC, Woolf BAR, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomization: the STROBE-MR Statement. JAMA 2021; 326: 1614-21.
12.
Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 2018; 362: k601.
13.
Zhao JH, Stacey D, Eriksson N, et al. Genetics of circulating inflammatory proteins identifies drivers of immune-mediated disease risk and therapeutic targets. Nat Immunol 2023; 24: 1540-51.
14.
Zhang Z, Cheng L, Ning D. Gut microbiota and sepsis: bidirectional Mendelian study and mediation analysis. Front Immunol 2023; 14: 1234924.
15.
Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018; 562: 203-9.
16.
Sun X, Liu B, Liu S, et al. Sleep disturbance and psychiatric disorders: a bidirectional Mendelian randomisation study. Epidemiol Psych Sci 2022; 31: e26.
17.
Palmer TM, Lawlor DA, Harbord RM, et al. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat Methods Med Res 2012; 21: 223-42.
18.
Kamat MA, Blackshaw JA, Young R, et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics 2019; 35: 4851-3.
19.
Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 2013; 37: 658-65.
20.
Greco MFD, Minelli C, Sheehan NA, et al. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat Med 2015; 34: 2926-40.
21.
Bowden J, Davey Smith G, Haycock PC, et al. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol 2016; 40: 304-14.
22.
Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015; 44: 512-25.
23.
Hotchkiss RS, Osmon SB, Chang KC, et al. Accelerated lymphocyte death in sepsis occurs by both the death receptor and mitochondrial pathways. J Immunol 2005; 174: 5110-8.
24.
Weber SU, Schewe J, Lehmann LE, et al. Induction of Bim and Bid gene expression during accelerated apoptosis in severe sepsis. Crit Care 2008; 12: R128.
25.
Pan J, Kunkel EJ, Gosslar U, et al. A novel chemokine ligand for CCR10 and CCR3 expressed by epithelial cells in mucosal tissues. J Immunol 2000; 165: 2943-9.
26.
Wuyts A, Haelens A, Proost P, et al. Identification of mouse granulocyte chemotactic protein-2 from fibroblasts and epithelial cells. Functional comparison with natural KC and macrophage inflammatory protein-2. J Immunol 1996; 157: 1736-43.
27.
Zweifel LS, Kuruvilla R, Ginty DD. Functions and mechanisms of retrograde neurotrophin signalling. Nature reviews. Neuroscience 2005; 6: 615-25.
28.
Costa HS, Lima MMO, Figueiredo PHS, et al. Prognostic value of serum brain-derived neurotrophic factor levels in patients with Chagas cardiomyopathy. Mem Inst Oswaldo Cruz 2018; 113: e180224.
29.
Bouralexis S, Findlay DM, Evdokiou A. Death to the bad guys: targeting cancer via Apo2L/TRAIL. Apoptosis 2005; 10: 35-51.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.