Introduction
Breast cancer – the most prevalent form of invasive malignancy – is the primary cause of cancer-related deaths among women due to its high incidence and mortality rates [1]. In 2020, breast cancer led to nearly 685,000 female fatalities globally and represented 30% of the anticipated cancer incidence in women for 2021 [2], underscoring the significant prevalence and mortality rates. Given the limited accessibility of breast cancer treatments and the costly nature of clinical trials [3], there is a critical need to investigate potential biomarkers linked to the development of breast cancer.
Breast cancer is influenced by a variety of both internal and external risk factors. Numerous epidemiological studies have identified mediators of breast cancer, with some Mendelian randomization (MR) studies confirming biomarkers associated with the disease. For instance, insulin-like growth factor-1 levels have been linked to a moderate increase in breast cancer risk [4]. Mitochondrial dysfunction, driven by genetic factors, has also been shown to play a causal role in breast cancer, with certain mitochondria-related genes implicated in disease development [5]. Additionally, serum C-reactive protein (CRP) has emerged as a potential biomarker for assessing overall cancer risk and risks specific to certain sites [6]. Despite these findings, research on the connection between the metabolome and breast cancer risk remains limited. Metabolomics, which focuses on the study of small molecules related to metabolic processes, can offer valuable insights when integrated with other histological platforms [7]. Understanding the causal relationships between metabolites and breast cancer development is crucial, as it may provide genetic evidence supporting the impact of key blood metabolites on breast cancer risk.
Genome-wide association studies (GWAS) are instrumental in identifying correlations across the genome between traits and single-nucleotide polymorphisms (SNPs), shedding light on the significance and impact of various genetic variants on different traits [8]. Recent GWAS have successfully identified causal links between the human metabolome and diseases [9]. On the other hand, MR analysis, a more robust method for inferring causality that has emerged in recent years, leverages genetic variation as instrumental variables (IV) to evaluate the causal relationship between risk factors and disease outcomes, thereby mitigating reverse causality bias [10]. Conducting a two-sample MR analysis necessitates data from distinct sources, such as two independent GWAS, to ascertain exposure and outcomes [11, 12]. In this study, we utilized two GWAS databases operating at different levels to perform a large-scale, two-sample MR analysis, systematically examining 100 human blood metabolites and pinpointing potential causal associations with breast cancer incidence.
Material and methods
Research design
This study utilized the publicly available GWAS database for a two-sample MR analysis to investigate the causal relationship between human blood metabolites and breast cancer. Ethical approval for data collection and written informed consent from participants were obtained in the original GWAS. SNPs were used as instrumental variables in this study, ensuring that they met the basic assumptions required for MR analysis.
Assumption 1: Genetic variants must be strongly associated with human blood metabolites.
Assumption 2: The genetic variants may be associated with the development of breast cancer specifically through human blood metabolites.
Assumption 3: Genetic factors must not be associated with any confounders of human blood metabolites and breast cancer.
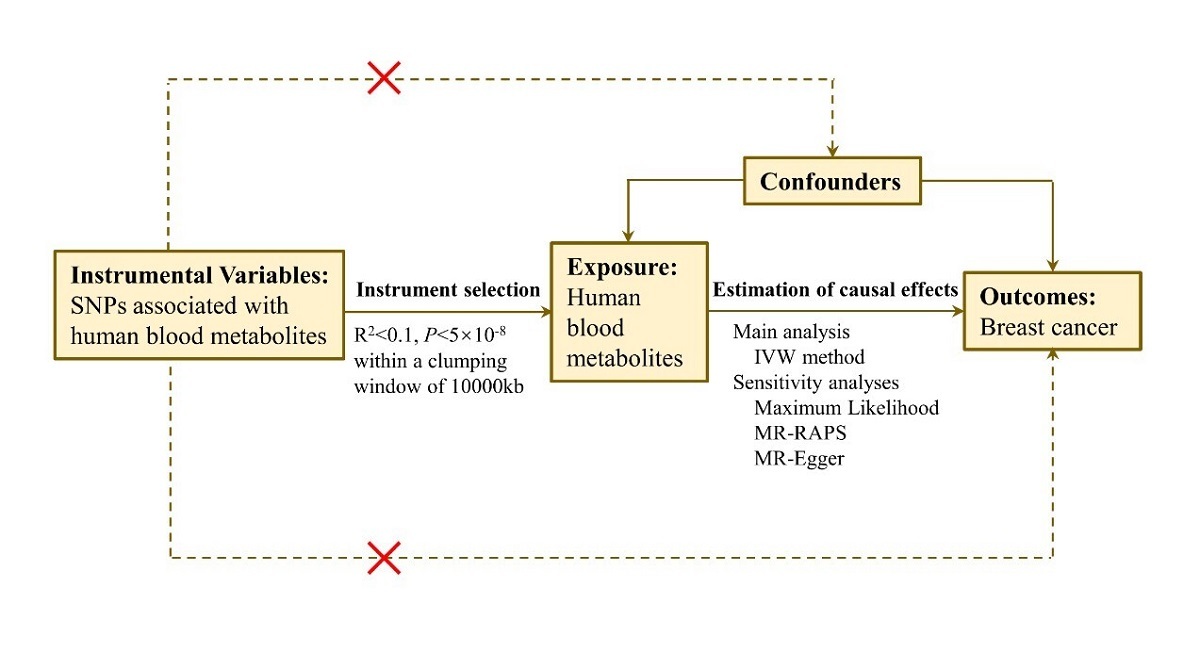
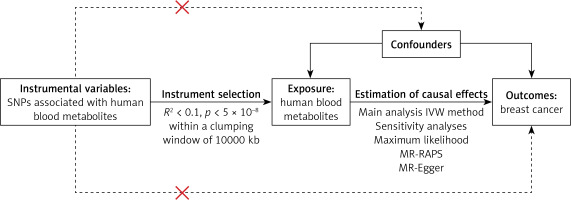
The study design process is illustrated in Figure 1. Approval for this study was obtained from the Affiliated Cancer Hospital of Harbin Medical University. Written informed consent was obtained from all participants or their legal representatives during the recruitment process.
Figure 1
Diagram of the Mendelian randomization (MR) study design. This MR study aimed to investigate the causal associations between human blood metabolites (exposure) and breast cancer (outcome). The assumption was that the instrumental variables are associated with metabolites, but not with confounders, and they influence the risk of breast cancer only through the association with metabolites, not confounders
SNP – single-nucleotide polymorphism, IVW – inverse-variance weighted, MR-RAPS – Mendelian randomization robust adjusted profile score.
Data sources
The study utilized human blood metabolite exposure and breast cancer genomic data from the Integrated Epidemiology Research Center’s Open Genome-Wide Association Studies (IEU OpenGWAS project) database, which includes two extensive GWAS cohorts totaling 265,554 individuals of European ancestry (detailed in Table I). The human blood metabolite data were sourced from Shin et al.’s study, analyzing 453 metabolic profiles of 7,824 participants with approximately 3 million SNPs. The outcome data came from a cohort study by Sakaue et al., involving 257,730 participants with 138,389 cases and 240,341 controls [13]. In this MR study, SNPs associated with 453 metabolites from the exposure cohort were examined to reflect blood metabolite expression at the gene level.
Table I
Associations between metabolites and risk of breast cancer in sensitivity analysis
Data processing
Exposure data screening
A total of 104 SNPs associated with the exposure cohort were extracted from the GWAS database based on the screening criteria of p < 5 × 10–8 [14]. To ensure independence among individual human blood metabolites, standard parameters for linkage disequilibrium removal were applied: linkage disequilibrium coefficient R2 < 0.1, with a window size of 10,000 kb. The strength of the selected SNPs was evaluated using the F-statistic with a window size of 10,000 kb, and SNPs with F > 10 was excluded.
Processing of outcome data
After merging the exposure data with the outcome data, the processed metabolites were aligned with the GWAS data on breast cancer incidence to pinpoint the instrumental variables linked to the outcome. Following this, the data sets were harmonized based on the statistical parameters of human blood metabolites and the GWAS data on breast cancer sharing the same loci, ensuring that the effect values of human blood metabolites and breast cancer were aligned to the same effect allele. Metabolites with less than three relevant SNPs in the genome were excluded, as at least three SNPs are required to be associated with exposure in certain MR sensitivity analyses [15]. Ultimately, 100 significant human blood metabolites were included in this study for further analysis.
Statistical analysis
The study primarily utilized the inverse variance weighting (IVW) method as the primary MR method to investigate the causal relationship between blood metabolite concentrations and breast cancer risk [16]. Cochran’s Q test was used, with a p-value greater than 0.05 indicating homogeneity in the results [17]. In cases of non-heterogeneous results, a fixed-effects model was employed, while a random-effects model was used for heterogeneous results to evaluate the MR effect. Sensitivity analyses were conducted to validate the reliability of IVW, including the maximum likelihood method, MR-robust adjusted profile scoring (MR-RAPS), and the MR-Egger method to detect horizontal pleiotropy [18]. Causal estimates between metabolites and breast cancer risk were presented as odds ratios (ORs) with corresponding 95% confidence intervals (CIs).
Data extraction, processing, and analysis were conducted using the TwoSampleMR (version 0.5.6) software package in R (version 4.1.3). A statistically significant difference was defined as p < 0.05.
Results
Results of instrumental variable screening
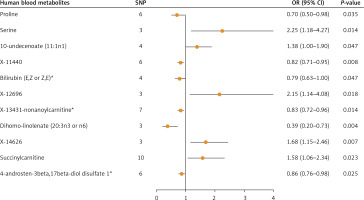
In this study, instrumental variables were carefully selected based on screening principles and criteria to identify 100 human blood metabolites associated with breast cancer. Cochran’s Q test was used to assess heterogeneity, with the random-effects model applied in the presence of heterogeneity and the fixed-effects model used when no heterogeneity was detected. The causal relationship between human blood metabolites and breast cancer risk was then examined using the IVW method. The analysis revealed that out of the 100 metabolites, 11 were found to have a causal link with breast cancer: proline, serine, 10-undecenoate (11:1n1), X-11440, bilirubin (E,Z or Z,E)*, X-12696, X-13431, X-13431-nonanoylcarnitine*, dihomo-linolenate (20:3n3 or n6), X-14626, succinylcarnitine, and 4-androsten-3beta,17beta-diol disulfate 1* (Figure 2). Detailed results can be seen in Supplementary Tables SI–SIII.
Associations of important human blood metabolites with breast cancer risk
The IVW method was utilized in the primary Mendelian randomization analysis to assess the causal relationships between 11 key human blood metabolites and the risk of breast cancer. The outcomes are comprehensively illustrated in Figure 2. The forest plot highlights five human blood metabolites that were identified as risk factors for breast cancer: serine (OR = 2.25; 95% CI: 1.18–4.27), 10-undecenoate (11:1n1) (OR = 1.38; 95% CI: 1.00–1.90), X-12696 (OR = 2.15; 95% CI: 1.14–4.08), X-14626 (OR = 1.68; 95% CI: 1.15–2.46), and succinyl carnitine (OR = 1.58; 95% CI: 1.06–2.34). Notably, serine and X-12696 exhibited the most robust associations with breast cancer risk, with ORs of 2.25 and 2.15, respectively.
Associations of the identified important human blood metabolites with breast cancer risk
The results of the analysis in Figure 3 indicate that alleles of five metabolites, including proline, X-11440, bilirubin (E,Z or Z,E)*, X-13431-nonanoylcarnitine*, and 4-androsten-3beta,17beta-diol disulfate 1*, are negatively associated with breast cancer risk, suggesting a decrease in risk with higher allele counts. Conversely, alleles of five blood metabolites, i.e. serine, 10-undecenoate (11:1n1), X-12696, X-14626, and succinylcarnitine, are positively linked to breast cancer risk, indicating an increase in risk with higher allele counts. These findings align with the primary MR analysis, which identifies serine, 10-undecenoate (11:1n1), X-12696, X-14626, and succinyl carnitine as potential biomarkers for breast cancer development.
Figure 3
Associations between genetic variants of identified metabolites and the risk of breast cancer. The line indicates the estimate of the causal effect using the inverse-variance weighted method. Circles indicate associations of each genetic variant related to metabolites with the risk of breast cancer. Error bars indicate 95% confidence interval. A – Proline; B – serine; C – 10-undecenoate (11:1n1); D – X-11440; E – bilirubin (E,Z or Z,E)*; F – X-12696; G – X-13431--nonanoylcarnitine*; H – dihomo-linolenate (20:3n3 or n6); I – X-14626; J – succinylcarnitine; K – 4-androsten-3beta, 17beta-diol disulfate 1*
SNP – single-nucleotide polymorphism.

Eleven important human metabolites identified were found to have 55 SNPs, with detailed characterization of each SNP variant provided in Supplementary Tables SIV–SXIV.
Sensitivity analysis
In sensitivity analyses using both the maximum likelihood and MR-RAPS methods, genetically determined serine, 10-undecenoate (11:1n1), X-12696, X-14626, and succinyl carnitine were found to be significantly associated with an increased risk of breast cancer development. MR-Egger regression intercept results indicated no evidence of directed pleiotropy among these five human blood metabolites and breast cancer risk. Therefore, these metabolites were identified as potential biomarkers for assessing breast cancer risk (Table I). Specifically, genetically determined serine (OR, 2.25; 95% CI: 1.18–4.27), 10-undecenoate (11:1n1) (OR = 1.38; 95% CI: 1.00–1.90), X-12696 (OR = 2.15; 95% CI: 1.14–4.08), X-14626 (OR = 1.68; 95% CI: 1.15–2.46), and succinyl carnitine (OR = 1.58; 95% CI: 1.06–2.34) showed an increased risk of breast cancer per 1 standard deviation increase.
Discussion
The combined metabolomics and genomics approach in this MR study offers novel insights into the risk of breast cancer and potential drug targets. Out of 100 human blood metabolites examined, five metabolites were found to have potential causal links with breast cancer: serine, 10-undecenoate (11:1n1), X-12696, X-14626, and succinylcarnitine. This indicates that genetically predicted higher levels of these metabolites may be linked to an increased risk of breast cancer.
Serine is a crucial precursor for the synthesis of various essential biomolecules such as proteins, lipids, nucleotides, and other amino acids, playing a central role in biosynthetic reactions necessary for cell division and growth [19]. The involvement of serine in cancer progression has garnered significant attention in the academic community. Research indicates that many cancer cells rely heavily on serine as a primary source of 1C units [20]. Previous studies have extensively validated the impact of serine on cancer development. For instance, oncogenes have been found to target enzymes in the serine biosynthetic pathway (SBP) [21], with the expression of these enzymes linked to inflammation in breast cancer. Additionally, elevated serine synthesis has been observed in breast cancer tissues [22]. Mechanisms through which increased serine synthesis accelerates carcinogenesis include altering glucose carbon flux, maintaining specific NAD(P)/NAD(P)H ratios, and regulating metabolite synthesis or expression [23, 24]. Building on these insights, our study further supports the role of serine as a mediator in breast cancer development, suggesting that targeting mitochondrial serine synthesis could be a promising strategy to impede breast carcinogenesis.
10-Undecenoate (11:1n1), a metabolite associated with gut microbiota, has been linked to various diseases. However, there is a lack of experimental evidence regarding its relationship with breast cancer risk or the impact of breast cancer on blood 10-undecenoate (11:1n1) levels. Further research is required to assess the role of blood 10-undecenoate (11:1n1) concentrations in diagnosing and treating breast cancer. Studies have suggested a potential causal link between 10-undecenoate (11:1n1) and Crohn’s disease, depression [25, 26], and low concentrations in patients with non-alcoholic fatty liver disease [27]. This neutral hydrophobic molecule remains poorly understood in the literature, but lifestyle factors such as diet and habits may influence breast cancer risk [28]. Overall, these findings offer insight into exploring the interplay of intestinal flora, metabolism, and breast cancer treatment, as well as shedding light on the connections between depression, Crohn’s disease, and breast cancer development.
X-12696 and X-14626 are newly discovered blood metabolites that have not been previously documented in the scientific literature. Interestingly, X-12696 has shown a strong association with breast cancer risk, ranking second only to serine. This highlights the importance of further research to investigate the role of X-12696 in the human body.
Succinyl carnitine, an acylcarnitine involved in fatty acid metabolism and mitochondrial function [29, 30], has been linked to various health conditions. Studies have shown elevated levels of succinyl carnitine in blood associated with Alzheimer’s disease and maternal concentrations during pregnancy, possibly contributing to coronary heart disease in offspring [31, 32]. Additionally, succinyl carnitine has been associated with total cholesterol, low-density lipoproteins, and breast cancer risk, highlighting its potential as a significant factor in disease development. Furthermore, as a newly identified urinary biomarker for γ-hydroxybutyric acid, a substance linked to brain metabolism and recreational drug use, succinyl carnitine has also been approved for treating narcolepsy [33]. These findings underscore the need for further research on the relationship between exogenous substances, e.g. recreational drugs and sleep disorder medications, and breast cancer risk. The study suggests potential associations between breast cancer and conditions such as coronary heart disease and Alzheimer’s disease, warranting further investigation for a better understanding of disease pathogenesis.
Our study has several strengths. Firstly, it is among the limited number of systematic MR studies that utilize blood metabolites as exposures to evaluate their causal impact on breast cancer risk. Secondly, this MR study utilized data from two extensive GWAS, enabling us to draw valid causal conclusions with robust statistical power. Thirdly, the study adhered to rigorous quality control measures and included a variety of sensitivity analyses and validity assessments, ensuring the stability and reliability of the results.
This study also has limitations. Firstly, the GWAS data used were solely from white European populations, thus limiting the generalizability of our findings to other racial and ethnic groups. Further research is necessary to confirm whether our results are applicable to other populations. Secondly, the lack of detailed demographic information, such as age and gender, in the extracted data prevented subgroup analyses from being conducted. In addition, due to constraints in time and funding, experimental validation was not performed.
In conclusion, this systematic meta-analysis identified serine, 10-undecenoate (11:1n1), X-12696, X-14626, and succinyl carnitine as potential biomarkers for predicting the risk of developing breast cancer. Specifically, serine and the previously unidentified blood metabolite X-12696 demonstrated the most significant associations with breast cancer prognosis.