Introduction
Otosclerosis (OTSC) is one of the major causes of progressive hearing loss (HL) in young adults. It results from pathological bone remodeling of the otic capsule. The new spongy bone forms typically around the footplate of the stapes, leading to its fixation, but the cochlea may also be involved. As movements of stapes are essential for the transfer of a sound wave to the inner ear, conductive or mixed hearing impairment develops. OTSC has an average age of onset in the third decade of life, but it may vary from the first to the sixth decade. It affects both sexes, with an approx. 1 : 2 female predominance. The prevalence of clinical OTSC in the Caucasian population is estimated at 0.3–0.4% and it is rather rare in Africans and Asians. Histological OTSC is found in up to 2.5% of cadaver temporal bones, which indicates that about 80% of OTSC is subclinical [1–3].
The etiology of OTSC is still poorly understood. It is considered to be complex, with the involvement of various environmental and genetic factors. Studies on the genetic background of OTSC distinguish between patients with a positive family history and sporadic cases. In familial cases the inheritance of OTSC is thought to be autosomal dominant with variable penetrance, but except for several delineated loci no causative variant has yet been identified [4]. For the more common sporadic OTSC cases, several population-based case-control studies have been performed to identify associations of different single nucleotide polymorphisms (SNPs) with OTSC [5–14]. Attempts to replicate the results often do not yield conclusive data. While in one population some of the variants are associated with OTSC, in another the hypothesis is rejected. This illustrates the obstacles to identify a major genetic factor associated with OTSC susceptibility.
It is of particular importance to search for genetic predisposing factors that could improve our understanding of complex disorders [15, 16] and advance precision medicine in the context of diagnosis and treatment options. To the best of our knowledge, there are no data on the genetic background of OTSC in patients originating from the central-eastern part of Europe. Here, we have focused on SNPs in TGFB1 (rs1800472) and RELN (rs39335, rs39350, rs39374) because they represent variants found to be significantly associated with OTSC in different populations and concurrently lack of their association with OTSC was reported in no more than one study. The research objective was to verify whether the selected SNPs are associated with surgically confirmed OTSC in Polish patients.
Material and methods
Patients’ data
A group of 94 patients (59 females and 35 males, mean age at HL diagnosis 36.3 years) was recruited from subjects with OTSC initially diagnosed using standard audiometric tests. All patients had no family history of OTSC and presented a characteristic air-bone gap in affected ears in pure-tone audiometry results. Diagnosis of OTSC was subsequently confirmed during stapedotomy (mean age at surgery 43.4 years). The control group consisted of 198 (111 females and 87 males) age-matched individuals with no family history of HL. All tested subjects gave informed consent for participation in the study in accordance with the tenets of the Declaration of Helsinki.
Genetic testing
Genomic DNA was isolated from whole blood samples or buccal swabs using a standard salting out procedure or automated method (Maxwell RSC Instrument, Promega, Germany), respectively. Three SNPs located in the RELN gene (rs39335, rs39350, rs39374) and one in the TGFB1 gene (rs1800472) were genotyped using Custom TaqMan SNP Genotyping Assays (Applied Biosystems, CA, USA) and real-time PCR (ViiA7, Thermo Fisher Scientific). Independent confirmation of genotypes in randomly selected samples was performed for TGFB1 by standard Sanger sequencing. Samples were amplified, labeled with the BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific) and sequenced on a 3500xL Genetic Analyzer (Applied Biosystems). Detailed Sanger sequencing data are available upon request.
Statistical analysis
Hardy-Weinberg (HW) equilibrium was calculated for every SNP in patient and control groups using the χ2 test. Association between genotypes and OTSC was evaluated for a dominant, co-dominant and recessive mode of inheritance. Differences in genotype and allele distributions among the tested groups were calculated with the χ2 or Fisher’s exact test with the significance level at p < 0.05. Odds ratios (ORs) with 95% confidence intervals (95% CI) were determined. All statistical analysis were performed using TIBCO Statistica 13 software (Tulsa, OK, USA). Non-random association of RELN rs39335, rs39350, rs39374 SNPs was tested by the linkage disequilibrium (LD) test and haplotype blocks were constructed based on genotype data [17, 18].
Results
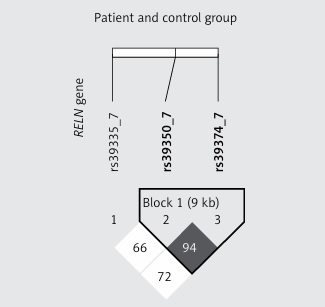
Data containing allele and genotype frequencies are listed in Table I. In both patient and control groups genotyping results of a particular SNP were in HW equilibrium. For every SNP the distribution of reference and alternative alleles between patient and control groups was similar and no statistically significant differences were observed. Testing the association between OTSC and RELN as well as TGFB1 genotypes under dominant, co-dominant or recessive models did not reveal statistically significant differences. In the tested subjects there were no sex-dependent differences in genotype and allele distributions (Table II). We found that RELN rs39350 and rs39374 form a haplotype (Figure 1) but none of the variant combinations, i.e. CA (χ2 = 0.64; p = 0.42), TG (χ2 = 0.01; p = 0.91) or CG (χ2 = 3.54; p = 0.06) for rs39350 and rs39374, respectively, was significantly associated with OTSC development. Genotypes obtained by Sanger sequencing were identical with those obtained by real-time PCR.
Table I
Distribution of RELN and TGFB1 genotypes and alleles in otosclerosis patients and controls
Table II
Sex-specific differences in allele frequency between otosclerosis patients and controls for the tested RELN and TGFB1 variants
Discussion
Here we tried to replicate previously reported findings of genetic associations in OTSC. Our study group comprised Polish patients with surgically confirmed OTSC as they represent a central-eastern European population for which no data on the role of genetic factors in OTSC have been available. For the investigation we selected variants in RELN and TGFB1 shown to be significantly and reproducibly associated with OTSC in different populations, but we did not confirm the initially described associations.
The TGFB1 gene encodes TGFB1, a multi-functional growth factor playing a role in proliferation, migration, differentiation and survival of many different cell types. In the skeletal system, it affects the function of both osteoclasts and osteoblasts and is considered the most important factor in maintaining the balance between bone resorption and deposition. The features make TGFB1 a good candidate gene for OTSC. A significant protective role of TGFB1 rs1800472 variant allele in OTSC development was demonstrated for Belgian-Dutch and French (OR = 0.423) [19], Tunisian (OR = 0.274) [20], Hungarian (OR = 0.267) [10] and British (OR = 0.51) [8] populations. In our cohort there was no statistically significant difference in the distribution of the TGFB1 variant between the patient and control group. Considering the low population frequency of the analyzed variant, a larger cohort would be needed to draw a reliable conclusion regarding the TGFB1 variant association with OTSC.
RELN encodes reelin, a glycoprotein involved in neuronal migration. Its relation to OTSC remains unclear but RELN expression was confirmed in the inner ear and in stapes footplates [12]. For RELN rs39335 a significant protective effect was found for Belgian-Dutch and French (OR = 0.741) [12], Tunisian (OR = 0.575) [14] and combined German, Italian, Swiss and Romanian (OR = 0.571) [13] populations. However, for Hungarian [10] and British [8] patients, similarly as in our patient group, no association with OTSC was observed.
The remaining two RELN SNPs, rs39350 and rs39374, were reported to increase the susceptibility to OTSC. A significant association was provided for Belgian-Dutch and French (OR = 1.367 and 1.441, respectively) [12], Tunisian (OR = 1.513 and 1.529, respectively) [14] and combined German, Italian, Swiss and Romanian (OR = 1.263 and 1.200, respectively) [13] populations. No evidence for an association between the two SNPs and OTSC was found for Hungarian patients or in the present study.
All studies mentioned above differed in the numbers of participants. In some of them the patient groups were more numerous, but in many of the studies the number of tested OTSC patients was around 100-150, which corresponds to our study group. To verify the quality of genotyping for selected samples a second method was applied. The results together with the data of statistical analysis showing that the control and patient groups are in HW equilibrium for every tested SNP suggest that the study was conducted correctly [21].
Our data show that none of the analyzed SNPs is associated with the susceptibility to OTSC in Polish patients. No association between RELN and OTSC was demonstrated for Hungarian and British patients either, but our study shows for the first time no association between TGFB1 and OTSC. The results underline again the complexity and high interpopulation variability of factors predisposing to OTSC development [1] and reinforce the suggestion that OTSC may represent not a single entity but a heterogenous group of diseases with different environmental and genetic factors involved.
In a recently conducted study division of patients into those with an OTSC positive and negative family history revealed different genetic associations. The strongest association among non-familial cases was observed for rs1800472 in TGFB1 [8]. Although our patients also represent sporadic OTSC cases, we did not replicate the finding. However, in the light of those results, it is more understandable why we did not detect any association for RELN SNPs as was found in patients with familial OTSC [8]. Despite the lack of genetic associations, negative results still provide valuable data guiding further research [22, 23]. In order to increase the probability of identifying an OTSC predisposing factor, it would be necessary to perform genome-wide association studies on a large, clinically homogeneous patient group.