Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Section Editors
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
NEPHROLOGY / CLINICAL RESEARCH
An improved random forest algorithm for tracing the origin of metastatic renal cancer tissues
1
Department of Urology, Chengdu Seventh People’s Hospital (Affiliated Cancer Hospital of Chengdu Medical College), Chengdu, China
2
Department of Imaging, West China Second Hospital, Sichuan University, Sichuan, China
Submission date: 2023-02-27
Final revision date: 2023-06-14
Acceptance date: 2023-06-28
Online publication date: 2023-07-11
Corresponding author
KEYWORDS
metastatic renal cancertissue origin tracingReliefFk_RFw algorithmdifferential expression analysisTCGA database
TOPICS
ABSTRACT
Introduction:
Tracing the histological origin of metastatic renal cancer (MRC) and locating the pathological root cause lead to precise treatment and improved prognosis.
Material and methods:
A total of 3336 patient cases with clear tissue origins from The Cancer Genome Atlas (TCGA) database were screened as experimental data material and feature selection was performed using the differential expression method; the random forest (RF) algorithm was improved to establish a medical retrospective heterogeneous filtered feature selection random forest weighted (ReliefFk_RFw) model to locate tissue origins.
Results:
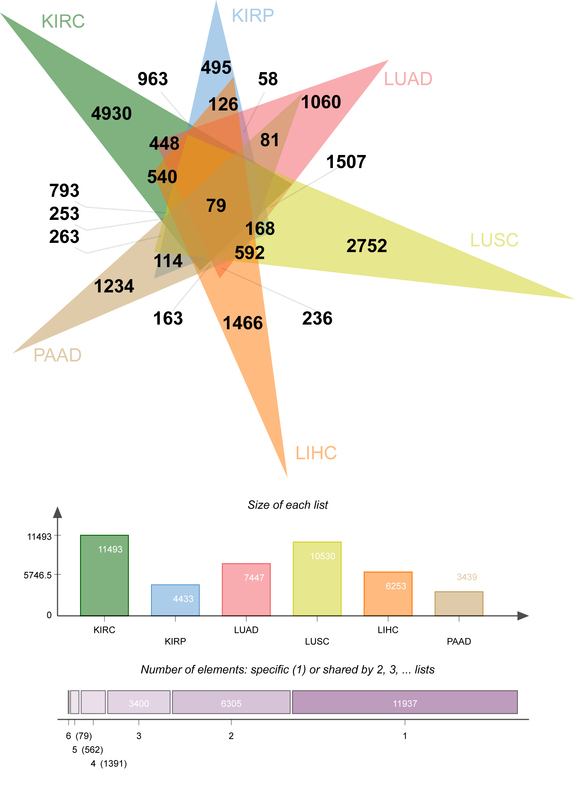
The differential expression analysis method screened 60 signature genes with good differential expression for tracing tissue origins (kidney renal clear cell carcinoma, kidney renal papillary cell carcinoma, lung adenocarcinoma, lung squamous cell carcinoma, liver hepatocellular carcinoma, pancreatic adenocarcinoma). Compared with traditional machine learning (support vector machine, decision tree, RF) models, the ReliefFk_RFw algorithm increased the average accuracy from 98.65%, 98.79% and 98.57% to 99.53%, the average precision from 95.58%, 96.40% and 96.54% to 99.36%, and the average sensitivity from 97.03%, 96.61% and 96.76% to 98.89%, mean specificity from 99.50%, 99.39% and 99.35% to 99.90%, and mean F1 score from 96.30%, 96.50% and 96.64% to 99.11%. The highest accuracy in localizing the origin of primary pancreatic cancer was achieved with 100.00% for different models of retrospective metrics.
Conclusions:
The improved ReliefFk_RFw model is best for comprehensive assessment and can be used to trace the origin of MRC tissue to assist physicians in diagnosis and treatment.
Tracing the histological origin of metastatic renal cancer (MRC) and locating the pathological root cause lead to precise treatment and improved prognosis.
Material and methods:
A total of 3336 patient cases with clear tissue origins from The Cancer Genome Atlas (TCGA) database were screened as experimental data material and feature selection was performed using the differential expression method; the random forest (RF) algorithm was improved to establish a medical retrospective heterogeneous filtered feature selection random forest weighted (ReliefFk_RFw) model to locate tissue origins.
Results:
The differential expression analysis method screened 60 signature genes with good differential expression for tracing tissue origins (kidney renal clear cell carcinoma, kidney renal papillary cell carcinoma, lung adenocarcinoma, lung squamous cell carcinoma, liver hepatocellular carcinoma, pancreatic adenocarcinoma). Compared with traditional machine learning (support vector machine, decision tree, RF) models, the ReliefFk_RFw algorithm increased the average accuracy from 98.65%, 98.79% and 98.57% to 99.53%, the average precision from 95.58%, 96.40% and 96.54% to 99.36%, and the average sensitivity from 97.03%, 96.61% and 96.76% to 98.89%, mean specificity from 99.50%, 99.39% and 99.35% to 99.90%, and mean F1 score from 96.30%, 96.50% and 96.64% to 99.11%. The highest accuracy in localizing the origin of primary pancreatic cancer was achieved with 100.00% for different models of retrospective metrics.
Conclusions:
The improved ReliefFk_RFw model is best for comprehensive assessment and can be used to trace the origin of MRC tissue to assist physicians in diagnosis and treatment.
REFERENCES (36)
1.
Bhanvadia RR, Baky FJ, Ashbrook CQ, et al. Pathologic fracture in metastatic kidney cancer: identifying widening disparities and opportunity for quality improvement. Urol Oncol 2022; 40: 384.e1-8.
2.
Rocca CJ, Cherqui S. Gene transfer to mouse kidney in vivo. Methods Mol Biol 2019; 1937: 227-34.
3.
Labriola MK, George DJ. Setting a new standard for long-term survival in metastatic kidney cancer. Cancer 2022; 128: 2058-60.
4.
Pal SK, Agarwal N. Kidney cancer: finding a niche for girentuximab in metastatic renal cell carcinoma. Nat Rev Urol 2016; 13: 442-3.
5.
Singh P, Agarwal N, Pal SK. Sequencing systemic therapies for metastatic kidney cancer. Curr Treat Options Oncol 2015; 16: 316.
6.
Afrit M, Yahyaoui Y, Bouzouita A, et al. Traitements médicaux des cancers du rein localement avancés et/ou métastatiques [Medical therapies for locally advanced/metastatic kidney cancer]. Presse Med 2015; 44: 135-43.
7.
Banyra O, Tarchynets M, Shulyak A. Renal cell carcinoma: how to hit the targets? Cent European J Urol 2014; 66: 394-404.
8.
Ouzaid I, Capitanio U, Staehler M, et al. Young academic urologists kidney cancer working group of the european association of urology. surgical metastasectomy in renal cell carcinoma: a systematic review. Eur Urol Oncol 2019; 2: 141-9.
9.
Zhong H, Li HY, Zhou T, et al. Rituximab therapy in adults with steroid-dependent nephrotic syndrome. Arch Med Sci 2023; 19: 577-85.
10.
Yuki M, Machida N, Sawano T, et al. Investigation of serum concentrations and immunohistochemical localization of 1-acid glycoprotein in tumor dogs. Vet Res Commun 2011; 35: 1-11.
11.
Solomon JP, Benayed R, Hechtman JF, et al. Identifying patients with NTRK fusion cancer. Ann Oncol 2019; 30 (Suppl_8): viii16-22.
12.
Kang X, Xia H, Skudder-Hill L, et al. Magnetic resonance imaging (MRI) and positron emission tomography (PET)/computed tomography features of atypical teratoid/rhabdoid tumors: case series and review. J Child Neurol 2022; 37: 1003-9.
13.
Lu D, Jiang J, Liu X, et al. Machine learning models to predict primary sites of metastatic cervical carcinoma from unknown primary. Front Genet 2020; 11: 614823.
14.
Wang Q, Xu Q, Chen J, et al. Establishment and evaluation of a novel molecular marker of tumor tissue origin. Chin J Cancer 2016; 26: 801-12.
15.
Zhao Y, Pan Z, Namburi S, et al. CUP-AI-Dx: a tool for inferring cancer tissue of origin and molecular subtype using RNA gene-expression data and artificial intelligence. Ebiomedicine 2020; 61: 103030.
16.
Carney JM, Kraynie AM, Roggli VL. Immunostaining in lung cancer for the clinician. Commonly used markers for differentiating primary and metastatic pulmonary tumors. Ann Am Thorac Soc 2015; 12: 429-35.
17.
Kalafi EY, Nor NAM, Taib NA, et al. Machine learning and deep learning approaches in breast cancer survival prediction using clinical data. Folia Biol (Praha) 2019; 65: 212-20.
18.
Park EY, Yi M, Kim HS, et al. A decision tree model for breast reconstruction of women with breast cancer: a mixed method approach. Int J Environ Res Public Health 2021; 18: 3579.
19.
Gadot R, Anand A, Lovin BD, et al. Predicting surgical decision-making in vestibular schwannoma using tree-based machine learning. Neurosurg Focus 2022; 52: E8.
20.
Zhou Y, Yu Z, Liu L, et al. Construction and evaluation of an integrated predictive model for chronic kidney disease based on the random forest and artificial neural network approaches. Biochem Biophys Res Commun 2022; 603: 21-8.
21.
Cheng X, Wang J, Zheng T, et al. Prediction of prognosis and survival of patients with gastric cancer by a weighted improved random forest model: an application of machine learning in medicine. Arch Med Sci 2022; 18: 1208-20.
22.
Gao R. Improvement of Random forest algorithm and its application in medical diagnosis system. Nanjing University of Posts and Telecommunications, 2020.
23.
Wang L, Liu S. Research on improved Random Forest Algorithm based on mixed sampling. J Nanjing Univ Posts Telecommun (Natural Science Edition) 2022; 01: 1673-5439.
24.
Yanping Xu. Urban air quality prediction model based on improved Random Forest algorithm: a case study of Chongqing Municipality. Chongqing Technology and Business University 2021.
25.
Attia ZI, Noseworthy PA, Lopez-Jimenez F, et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet 2019; 394: 861-7.
26.
Dong L, He W, Zhang R, et al. Artificial intelligence for screening of multiple retinal and optic nerve diseases. JAMA Netw Open 2022; 5: e229960.
27.
Choi E, Kim D, Lee JY, et al. Artificial intelligence in detecting temporomandibular joint osteoarthritis on orthopantomogram. Sci Rep 2021; 11: 10246.
28.
Zhu S, Lu B, Wang C, et al. Screening of common retinal diseases using six-category models based on efficientnet. Front Med (Lausanne) 2022; 9: 808402.
29.
Zheng X, Huang R, Liu G, et al. Development and verification of a predictive nomogram to evaluate the risk of complicating ventricular tachyarrhythmia after acute myocardial infarction during hospitalization: a retrospective analysis. Am J Emerg Med 2021; 46: 462-8.
30.
Cheng Q, Chen X, Wu H, et al. Three hematologic/immune system-specific expressed genes are considered as the potential biomarkers for the diagnosis of early rheumatoid arthritis through bioinformatics analysis. J Transl Med 2021; 19: 18.
31.
Jin C, Jiang Y, Yu H, et al. Deep learning analysis of the primary tumour and the prediction of lymph node metastases in gastric cancer. Br J Surg 2021; 108: 542-9.
32.
Doppalapudi S, Qiu RG, Badr Y. Lung cancer survival period prediction and understanding: deep learning approaches. Int J Med Inform 2021; 148: 104371.
33.
Liu H, Chen Y, Zhang Y, et al. A deep learning model integrating mammography and clinical factors facilitates the malignancy prediction of BI-RADS 4 microcalcifications in breast cancer screening. Eur Radiol 2021; 31: 5902-12.
34.
Tan WCC, Nerurkar SN, Cai HY, et al. Overview of multiplex immunohistochemistry/immunofluorescence techniques in the era of cancer immunotherapy. Cancer Commun (Lond) 2020; 40: 135-53.
35.
Tian M, Wang T, Wang P. Development and clinical validation of a seven-gene prognostic signature based on multiple machine learning algorithms in kidney cancer. Cell Transplant 2021; 30: 963689720969176.
36.
Taskesen E, Huisman SM, Mahfouz A, et al. Pan-cancer subtyping in a 2D-map shows substructures that are driven by specific combinations of molecular characteristics. Sci Rep 2016; 6: 24949.
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.