Current issue
Archive
Manuscripts accepted
About the Journal
Editorial office
Editorial board
Section Editors
Abstracting and indexing
Subscription
Contact
Ethical standards and procedures
Most read articles
Instructions for authors
Article Processing Charge (APC)
Regulations of paying article processing charge (APC)
CLINICAL RESEARCH
An immune escape-related gene signature for predicting the prognostic and immune microenvironment in osteosarcoma
1
Graduate School of Kunming Medical University, Chenggong District, Kunming, Yunnan, China
2
920th Hospital of Joint Logistics Support Force of Chinese People’s Liberation Army, Xishan District, Kunming, Yunnan, China
3
Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan, China
Submission date: 2024-05-22
Final revision date: 2024-11-18
Acceptance date: 2025-01-05
Online publication date: 2025-04-08
Corresponding author
Yongqing Xu
920th Hospital of Joint Logistics Support Force of Chinese People’s Liberation Army, 212 Daguan Road Xishan District, Kunming Yunnan, China
920th Hospital of Joint Logistics Support Force of Chinese People’s Liberation Army, 212 Daguan Road Xishan District, Kunming Yunnan, China
Article (PDF)
Supplementary files
References (27)
An immune escape-related - Supplementary figs.pdf
An immune escape-related - Supplementary Table SI.XLSX
An immune escape-related - Supplementary Table SII.XLSX
An immune escape-related - Supplementary Table SIII.XLSX
An immune escape-related - Supplementary Table SIV.XLSX
An immune escape-related - Supplementary Table SV.XLSX
An immune escape-related - Supplementary Table SVI.XLSX
KEYWORDS
TOPICS
ABSTRACT
Introduction:
This study aimed to develop an immune escape-related gene signature for prognostic prediction and clarification of the immune microenvironment in osteosarcoma, a predominant malignant bone tumor in pediatric and adolescent populations.
Material and methods:
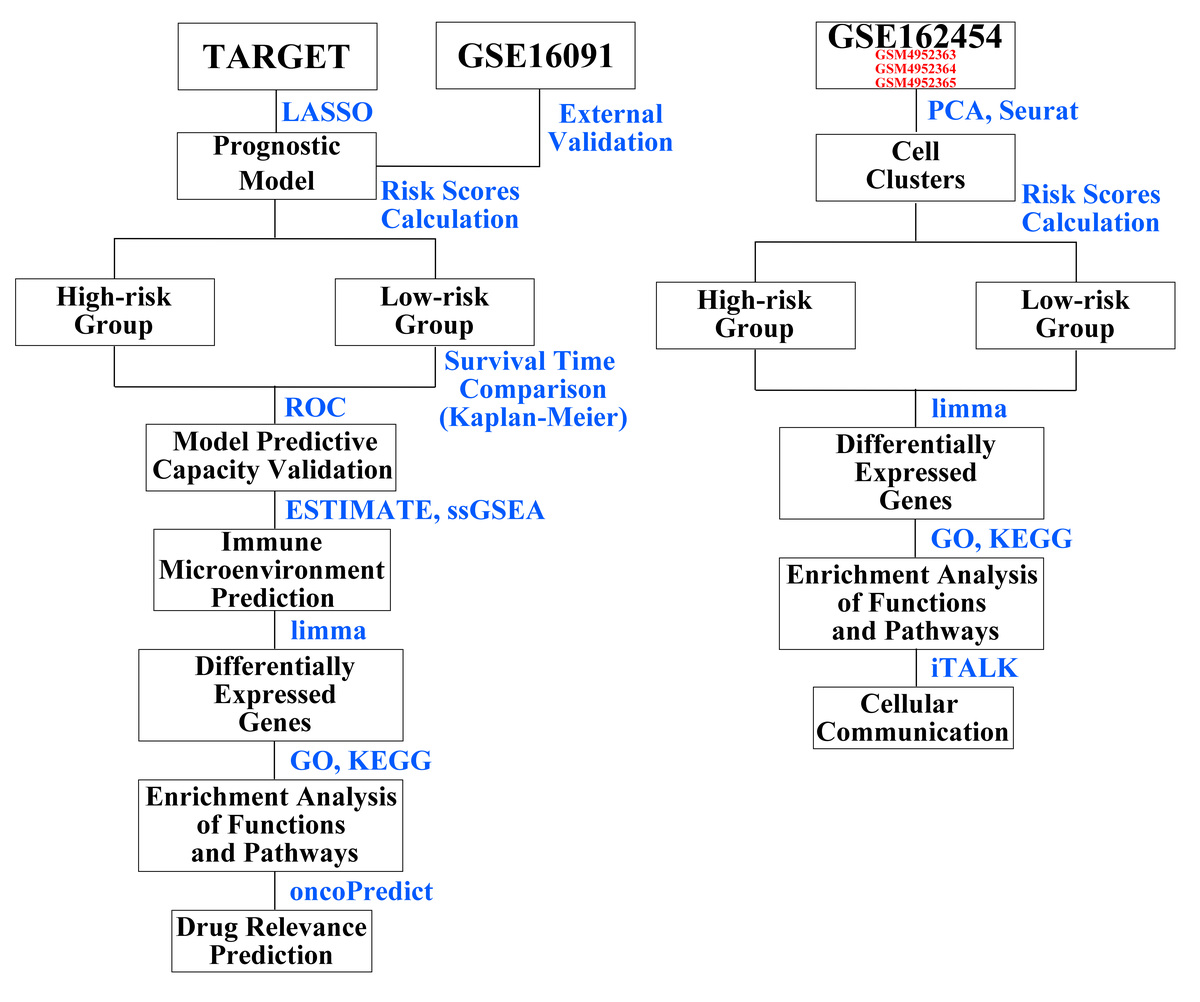
This study used transcriptomic and genomic data from various databases (Therapeutically Applicable Research to Generate Effective Treatments and Gene Expression Omnibus). A prognostic model was established using the least absolute shrinkage and selection operator method, followed by rigorous statistical analysis. Additionally, the study involved the investigation of differential pathways and single-cell data analysis to understand the immune escape mechanisms in osteosarcoma.
Results:
The study successfully developed an immune escape-related gene model that stratifies patients with osteosarcoma into different prognostic groups with significant survival differences. It indicated that higher immune escape-related gene scores were associated with poor survival outcomes. Additionally, the model demonstrated efficacy in predicting the complexity and variability of the immune microenvironment in osteosarcoma, correlating with different immune cell infiltrations and immunotherapy responses. Furthermore, single-cell analysis revealed distinct molecular signatures and pathways associated with immune escape, emphasizing potential therapeutic targets in osteosarcoma management.
Conclusions:
The immune escape-related gene model provides a novel approach to understanding and predicting osteosarcoma prognosis. This model serves as a valuable tool for determining potential therapeutic targets and developing personalized treatment strategies. It emphasizes the importance of immune escape mechanisms in osteosarcoma progression and treatment.
This study aimed to develop an immune escape-related gene signature for prognostic prediction and clarification of the immune microenvironment in osteosarcoma, a predominant malignant bone tumor in pediatric and adolescent populations.
Material and methods:
This study used transcriptomic and genomic data from various databases (Therapeutically Applicable Research to Generate Effective Treatments and Gene Expression Omnibus). A prognostic model was established using the least absolute shrinkage and selection operator method, followed by rigorous statistical analysis. Additionally, the study involved the investigation of differential pathways and single-cell data analysis to understand the immune escape mechanisms in osteosarcoma.
Results:
The study successfully developed an immune escape-related gene model that stratifies patients with osteosarcoma into different prognostic groups with significant survival differences. It indicated that higher immune escape-related gene scores were associated with poor survival outcomes. Additionally, the model demonstrated efficacy in predicting the complexity and variability of the immune microenvironment in osteosarcoma, correlating with different immune cell infiltrations and immunotherapy responses. Furthermore, single-cell analysis revealed distinct molecular signatures and pathways associated with immune escape, emphasizing potential therapeutic targets in osteosarcoma management.
Conclusions:
The immune escape-related gene model provides a novel approach to understanding and predicting osteosarcoma prognosis. This model serves as a valuable tool for determining potential therapeutic targets and developing personalized treatment strategies. It emphasizes the importance of immune escape mechanisms in osteosarcoma progression and treatment.
REFERENCES (27)
1.
Niu J, Yan T, Guo W, et al. Identification of potential therapeutic targets and immune cell infiltration characteristics in osteosarcoma using bioinformatics strategy. Front Oncol 2020; 10: 1628.
2.
Redondo A, Cruz J, Lopez-Pousa A, Barón FJC, SEOM. SEOM clinical guidelines for the treatment of osteosarcoma in adults-2013. Clin Transl Oncol 2013; 15: 1037-43.
3.
Xie QK, Zhao YJ, Pan T, et al. Programmed death ligand 1 as an indicator of pre-existing adaptive immune responses in human hepatocellular carcinoma. Oncoimmunology 2016; 5: e1181252.
4.
Wildes TJ, Dyson KA, Francis C, et al. Immune escape after adoptive T-cell therapy for malignant gliomas. Clin Cancer Res 2020; 26: 5689-700.
5.
Chen Y, Xu J, Wu X, et al. CD147 regulates antitumor CD8+ T-cell responses to facilitate tumor-immune escape. Cell Mol Immunol 2021; 18: 1995-2009.
6.
Lu C, Klement JD, Smith AD, et al. p50 suppresses cytotoxic T lymphocyte effector function to regulate tumor immune escape and response to immunotherapy. J Immunother Cancer 2020; 8: e001365.
7.
Yu L, Zhang J, Li YJ. Effects of microenvironment in osteosarcoma on chemoresistance and the promise of immunotherapy as an osteosarcoma therapeutic modality. Front Immunol 2022; 13: 871076.
8.
De Jaeghere EA, Denys HG, De Wever O. Fibroblasts fuel immune escape in the tumor microenvironment. Trends Cancer 2019; 5: 704-23.
9.
Zhou Y, Yang D, Yang QC, et al. Single-cell RNA-seq reveals identity and heterogeneity of malignant osteoblast cells and TME in osteosarcoma. bioRxiv 2020; 2020.04. 16.044370.
10.
Fan TM, Roberts RD, Lizardo MM. Understanding and modeling metastasis biology to improve therapeutic strategies for combating osteosarcoma progression. Front Oncol 2020; 10: 13.
11.
Grünewald TG, Alonso M, Avnet S, et al. Sarcoma treatment in the era of molecular medicine. EMBO Mol Med 2020; 12: e11131.
12.
Gill J, Gorlick R. Advancing therapy for osteosarcoma. Nat Rev Clin Oncol 2021; 18: 609-24.
13.
Lu Y, Zhang J, Chen Y, et al. Novel immunotherapies for osteosarcoma. Front Oncol 2022; 12: 830546.
14.
Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43: e47.
15.
Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013; 4: 2612.
16.
Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell 2021; 184: 3573-87.e29.
17.
Wang Y, Wang R, Zhang S, et al. iTALK: an R package to characterize and illustrate intercellular communication. bioRxiv 2019:507871.
18.
Maeser D, Gruener RF, Huang RS. oncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief Bioinform 2021; 22: bbab260.
19.
Zhu XG, Chudnovskiy A, Baudrier L, et al. Functional genomics in vivo reveal metabolic dependencies of pancreatic cancer cells. Cell Metab 2021; 33: 211-21.e6.
20.
Chavez-Dominguez R, Perez-Medina M, Lopez-Gonzalez JS, Galicia-Velasco M, Aguilar-Cazares D. The double-edge sword of autophagy in cancer: from tumor suppression to pro-tumor activity. Front Oncol 2020; 10: 578418.
21.
Lu J, Tang H, Chen L, et al. Association of survivin positive circulating tumor cell levels with immune escape and prognosis of osteosarcoma. J Cancer Res Clin Oncol 2023; 149: 13741-51.
22.
Guo J, Tang H, Huang P, et al. Single-cell profiling of tumor microenvironment heterogeneity in osteosarcoma identifies a highly invasive subcluster for predicting prognosis. Front Oncol 2022; 12: 732862.
23.
Lei X, Lei Y, Li JK, et al. Immune cells within the tumor microenvironment: biological functions and roles in cancer immunotherapy. Cancer Lett 2020; 470: 126-33.
24.
Liu W, Hu H, Shao Z, et al. Characterizing the tumor microenvironment at the single-cell level reveals a novel immune evasion mechanism in osteosarcoma. Bone Res 2023; 11: 4.
25.
Liu Y, Feng W, Dai Y, et al. Single-cell transcriptomics reveals the complexity of the tumor microenvironment of treatment-naive osteosarcoma. Front Oncol 2021; 11: 709210.
26.
Griffin KH, Thorpe SW, Sebastian A, et al. Engineered bone marrow as a clinically relevant ex vivo model for primary bone cancer research and drug screening. Proc Natl Acad Sci USA 2023; 120: e2302101120.
27.
Cheng D, Zhang Z, Mi Z, et al. Deciphering the heterogeneity and immunosuppressive function of regulatory T cells in osteosarcoma using single-cell RNA transcriptome. Comput Biol Med 2023; 165: 107417.
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.